BAPG Fall 2025
Bay Area Population Genomics Conference @ Stanford
BAPG 2025 was a blast. Looking forward to seeing folks in Davis in Spring 2026! bapg2025.github.io/bapg2025stan...
07.12.2025 03:58 — 👍 8 🔁 6 💬 0 📌 0
BAPG Fall 2025
Bay Area Population Genomics Conference @ Stanford
Super excited about our schedule for BAPG at stanford on Dec 6. bapg2025.github.io/bapg2025stan... Amazing talks, a fabulous keynote, a lively poster session. A brilliant and interactive community. What’s not to love? 1/2 @sophiejwalton.bsky.social
24.11.2025 22:54 — 👍 14 🔁 7 💬 1 📌 1

Registration for BAPG 2025, Stanford Dec 6 2025
Talk submissions are now closed, but poster submissions are still open.
Please note that we already have 130+ people signed up and we will limit the sign up at 160. Don’t dawdle If you want to attend. Registration is free but required. forms.gle/M5BSyAaeUggn...
24.11.2025 22:54 — 👍 2 🔁 2 💬 0 📌 0
We have 115 people signed up already for BAPG! Great representation across career stages too. If you want to attend don't forget to sign up. It is free but required. @sophiejwalton.bsky.social and I are looking through the wonderful talk submissions and will announce the talks shortly. Stay tuned!
20.11.2025 18:29 — 👍 8 🔁 5 💬 0 📌 0

Dynamics of dN/dS within recombining bacterial populations
The ratio of nonsynonymous to synonymous substitutions (dN/dS) encodes important information about the selection pressures acting on protein-coding genes. In bacterial populations, dN/dS often decline...
The first is from former PhD student Zhiru Liu @zzzhiru.bsky.social (now in @bengrbm.bsky.social's group @ MSK) examining the long-term patterns of selective constraint – measured by the classical ratio of nonsynonymous to synonymous mutations (dN/dS) – within recombining populations of bacteria.
16.11.2025 15:26 — 👍 16 🔁 9 💬 2 📌 0
BAPG Fall 2025
Bay Area Population Genomics Conference @ Stanford
don't forget to register for BAPG at Stanford on Dec 6! @petrovadmitri.bsky.social and CEHG are hosting. talk submissions close on Nov 16 - bapg2025.github.io/bapg2025stan...
docs.google.com/forms/d/e/1F...
13.11.2025 18:55 — 👍 2 🔁 1 💬 0 📌 1
Beyond excited to share my PhD work thus far, now up as a preprint! We found that a transposable element insertion is responsible for the recent evolution of an novel color trait. Feeling thankful to everyone who has helped in this project and thrilled to continue learning about "sparkle"!
12.11.2025 22:58 — 👍 34 🔁 5 💬 2 📌 0
i will never stop laughing
12.11.2025 22:24 — 👍 2 🔁 0 💬 0 📌 0
Thank you!
12.11.2025 16:05 — 👍 0 🔁 0 💬 0 📌 0
And of course thank you to @benjaminhgood.bsky.social and @petrovadmitri.bsky.social for mentoring me through this adventure. n/n
11.11.2025 17:14 — 👍 3 🔁 1 💬 0 📌 0
Thanks to all my amazing collaborators! @ksxue.bsky.social , Jonas and Richa were instrumental to setting up this project! Also, huge shout out to Qing, @hgellert.bsky.social and Chih-Fu who contributed to the analysis and experiments. (20/n)
11.11.2025 17:14 — 👍 3 🔁 0 💬 1 📌 0
This suggests that conspecific strains can behave more like ecological species when competing within larger communities, even when their genomes appear to evolve as a single biological species. (19/n)
11.11.2025 17:14 — 👍 2 🔁 0 💬 1 📌 0
Together, our results illustrate that selection on conspecific strains in large communities is strong, but attenuates as communities stabilize in a manner reminiscent of negative frequency dependent selection. (18/n)
11.11.2025 17:14 — 👍 3 🔁 0 💬 1 📌 0
There are additional analyses in the paper that I didn’t cover here, including looking at how selection shifts across nutrient environments! (17/n)
11.11.2025 17:14 — 👍 3 🔁 0 💬 1 📌 0

Alt text: Data showing community cohesion arranged as a table. Rows are collision types, columns are species. Data shows that winners from both hosts are present in most collisions.
We also examined whether strain-level competition outcomes were correlated across species. Interestingly, we found little evidence for strain-level cohesion during coalescence, despite prior co-selection during community assembly and within each host. (16/n)
11.11.2025 17:14 — 👍 2 🔁 0 💬 1 📌 0

Alt text: Number of species over passages in community mixtures and control communities. Number of species in both cases equilibrates to ~50 species.
Our observations of strain-level coexistence were surprising because community mixtures did not have elevated diversity compared to autologous controls! (14/n)
11.11.2025 17:14 — 👍 2 🔁 0 💬 1 📌 0
Since the abiotic environment was held constant, we attributed these shifts in selection to biotic interactions within the community. This suggests that conspecific strains can coexist through niche partitioning in species rich communities. (13/n)
11.11.2025 17:14 — 👍 3 🔁 0 💬 1 📌 0

Alt text: Two panels. Top- An example of time varying selection for an f. plautii competition. Under constant selection, the one strain is expected to go extinct. However, selection attenuates and the strains continue to coexist. Bottom- Correlation between predicted strain diversity (f x (1-f)) and actual strain diversity. In most cases, observed strain diversity exceeds predicted strain diversity.
However, selection shifted at later generations in many competitions! Moreover, these shifts also tended to be biased towards maintaining coexistence. (12/n)
11.11.2025 17:14 — 👍 2 🔁 0 💬 1 📌 0

Alt text: Fig panel showing the distribution of early phase fitness (between passages 1 and 2) across all pairwise competitions.
We then estimated the time-varying relative fitnesses of conspecific strains. We found that selection during earlier generations was quite strong and predicted that one strain would dominate in most competitions. (11/n)
11.11.2025 17:14 — 👍 4 🔁 0 💬 1 📌 0

Alt text: Fig panel showing Bacteroides dorei strain dynamics across 4 replicate competitions. Annotations show that sampling error is small and the communities undergo large shifts in frequency. Dynamics across replicates are correlated. Empirical cumulative distributions are also shown of the distribution of frequency shifts due to sampling error, between replicate collisions and between timepoint 0 and 7.
We found that the dynamics of competing strains were characterized by large and deterministic shifts. These shifts exceeded what could be generated by measurement noise or drift, demonstrating that these dynamics were driven by strong selection. (10/n)
11.11.2025 17:14 — 👍 3 🔁 0 💬 1 📌 0

Alt text: Fig describing community coalescence. Two stabilized communities derived from different hosts are mixed in equal ratio to create a third community. The community that results from collision is passaged for 7 passages. Additional panel showing Marker SNVs for Escherichia coli is shown. Groups of correlated SNVs show that the E. coli strains from different hosts undergo strong selection.
We then monitored the dynamics of competing conspecific strains from each host through the course of the coalescence through shotgun metagenomics. This approach allowed us to follow pairwise strain competitions across many species in parallel! (9/n)
11.11.2025 17:14 — 👍 2 🔁 0 💬 1 📌 0

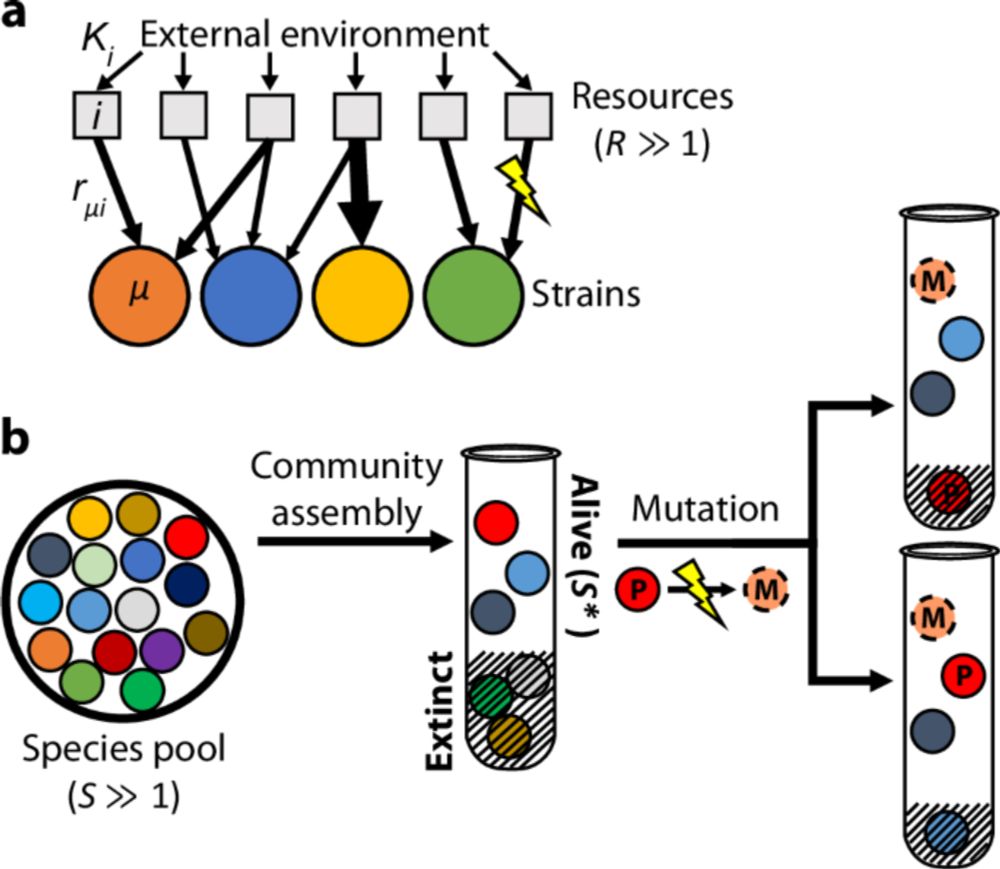
Alt text: cartoon describing how communities from fecal samples are derived. Stool from a healthy human donor is grown in rich media (mBHI or mGAM) w/ passages every 48 hrs. Communities are sequenced via metagenomic sequencing. Relative abundance bar plots are shown of community species composition over 5 passages (38 generations). Communities have many prominent gut families, including bacteroidaceae. Number of species over time is also shown. Communities stabilize to ~50 species.
We collected fecal samples from donors in the Bay Area and assembled in vitro gut communities from these samples in anaerobic conditions. Communities from different hosts shared some species, but harbored diverged strains of those species. (8/n)
11.11.2025 17:14 — 👍 2 🔁 0 💬 1 📌 0

Alt text: Fig panel from Li et al 2016 showing that donor and recipient strains in an FMT can coexist. Results are displayed as a table with species as rows and FMT recipients as columns. The outcomes of the FMT vary at both a species and subject level.
Microbial community coalescence experiments have been used to gain a lot of intuition on species-level selection. We were also inspired by strain level data that had been obtained from FMTs in therapeutic contexts (fig from DOI: 10.1126/science.aad8852). (7/n)
11.11.2025 17:14 — 👍 2 🔁 0 💬 1 📌 0

Alt text: Cartoon of community coalescence. Communities from two hosts are combined to create a third community.
It’s hard to distinguish between these models in natural microbiomes, so turned to lab systems to get some intuition. Specifically, we performed whole community competitions, or community coalescence, with in vitro gut communities derived from different human donors. (6/n)
11.11.2025 17:14 — 👍 3 🔁 0 💬 1 📌 0

Alt text: Muller plot of three species fluctuating in abundance over time. A new strain of one of the species invades from a global population and competes with the old strain.
Others have proposed that conspecific strains may compete as ‘ecological species’ and coexist via niche partitioning because we see lots of examples of multiple conspecific strains persisting in the same microbiome. (5/n)
11.11.2025 17:14 — 👍 2 🔁 0 💬 1 📌 0
However, some ecology theory suggests that diverse communities could select for suites of strains with small fitness differences within a community. DOI: 10.1016/j.tree.2006.08.003(4/n)
11.11.2025 17:14 — 👍 2 🔁 0 💬 1 📌 0
Microbiologists have shown us that conspecific strains can have large differences in important traits, such as carbohydrate metabolism. This could mean that conspecific strains have large fitness differences in any local environment (3/n).
11.11.2025 17:14 — 👍 2 🔁 0 💬 1 📌 0

Alt text: Cartoon phylogeny of 4 human hosts and their microbial strains. Arrows indicate that these strains are separate by ~100kya of evolution. ~10k SNVs separate the microbial strains.
Complex microbial communities, like the human gut microbiome, can harbor extensive strain-level diversity, with strains of the same species from different hosts differing by ~10k mutations. However, the fitness consequences of this diversity in community contexts are not well understood. (2/n)
11.11.2025 17:14 — 👍 6 🔁 1 💬 1 📌 0
Ph.D. Candidate working on maize. Chaotically interdisciplinary, but generally in the realm of pop gen, domestication, plants, and people.
From Delco & Philly (go birds! 🦅), PhD in progress at UC Davis 🐄
Fungi 🍄 | Plants 🌵 | Epigenetics 🧬 | Eco-Evo-Devo 🌀 | PhD Student, Stanford 🌲(he/him/il) #FirstGen #QueerinSTEM 🏳️🌈 ♾️
🌎 Science is for everyone.
🍄🟫 Views are my own.
Postdoc in Pritchard lab at Stanford University. Previous PhD co-advised by Yun Song and Rasmus Nielsen at UC Berkeley.
PhD candidate in Dmitri Petrov's lab @ Stanford
Evolution of social complexity & evolutionary genetics of social insects. Assoc Prof at Arizona State University
Assistant Prof at Case Western studying microbial interactions in the oral and vaginal microbiomes (and beyond!). Also find me outside, baking, coloring, or sleeping.
thelewinlab.com
The Cress Lab | Innovative Genomics Institute @ UC Berkeley | Microbiome Editing | Microbiome Delivery Technologies | Phage and MGE Functional Genomics | Hiring!
https://www.cresslab.bio/
Postdoc at @ETH_en 👩🏽🔬👩🏽💻 🧐
Microbial evolution | Human microbiome ecology | Infectious disease epidemiology
Postdoc at UC Berkeley | evolutionary genetics & genomics, plant mating systems, hybridization
PhD Student at UMich Statistics.
The account mostly trashes about urban planning and infrastructure.
Probability, Statistics, and Evolutionary Biology.
https://hanbin973.github.io
Scientist Human Gut microbiome @MIB_WUR 🇳🇱 - @DanoneResearch 🇨🇵 - now @RaesLab 🇧🇪 (@VIB_microbes @KU_Leuven). Opinions are my own.
/dee-AA-nah/|Postdoc at UCLA|computational biology PhD / popgen, genomics and conservation | she/her/ella 🇲🇽
PhD student in Fordyce & Petrov labs at Stanford | biophysics of evolutionary adaptation in proteins | using population genomics, synthetic biology, microfluidics
Evolutionary biologist, computational biologist, statistician. I like to develop mathematical models of evolutionary process and see how they fit to data. I also like cities where building apartments is legal.
Assistant Professor at University of Calgary, Canada | Understanding and editing microbiomes using ecology | Mountain and trail runner🏃♂️⛰ | Postdoc in 🇬🇧, PhD in 🇨🇭 (he/him)
https://erikbakkeren.com/
Principal Researcher in BioML at Microsoft Research. He/him/他. 🇹🇼 yangkky.github.io
Assistant professor at UNH, microbes, plants, ecology, evolution
Physics PhD student @ UF
Aspiring Biophysicist :)
I study sexual selection and speciation using theoretical and empirical approaches. Currently a Miller postdoctoral fellow at UC Berkeley.
PhD from UCSC '24.
BS from UVA '18 Biol + CS.
Website: https://kustra-matt.github.io/