
Kim Lab at UC Irvine
Visit the post for more.
The Kim Lab (sskimlab.org) at UC Irvine is recruiting motivated grad students interested in uncovering how transcriptional regulators direct cell fates. We accept students through the CMB (cmb.uci.edu) and MCSB (ccbs.uci.edu/education/mcsb/) PhD programs. Please share with prospective grad students!
30.10.2025 15:37 — 👍 9 🔁 8 💬 0 📌 0

More than 30% of this century’s science Nobel prizewinners immigrated: see their journeys
The most common destination for eventual Nobel laureates in physics, chemistry and medicine since 2000 is the United States, Nature has found.
Less than 70% of science Nobel prize winners awarded this century hail from the country in which they were awarded their prize.
“Mobility benefits everyone. Each newcomer brings fresh ideas, new techniques & different ways of looking at old problems”
🧪 #academicSky
www.nature.com/articles/d41...
09.10.2025 22:58 — 👍 104 🔁 42 💬 3 📌 4
ScienceDirect.com | Science, health and medical journals, full text articles and books.
Very happy that the review article by @magnushaughey.bsky.social and Imran Noorani on the evolutionary role of extra-chromosomal DNA in cancers just came out.
authors.elsevier.com/sd/article/S...
09.07.2025 13:43 — 👍 3 🔁 1 💬 0 📌 0

Decoupling chromatin hubs from gene control
Nature Structural & Molecular Biology - Chromatin hubs, which are formed during cell differentiation, are characterized by simultaneous interactions between multiple regulatory elements and...
🚨The formation of 3D chromatin hubs is a poorly understood
phenomenon.
I wrote a N&V about the excellent work Karpinska, Zhu and colleagues, from @mariekeoudelaar.bsky.social lab, who shed new light on the formation and function of these structures.
rdcu.be/elPt3
13.05.2025 10:13 — 👍 50 🔁 18 💬 3 📌 0
Thanks for reading and we would be delighted to hear your comments/suggestions.
22.05.2025 17:37 — 👍 1 🔁 0 💬 0 📌 0
I’m also incredibly proud of our multi-institutional, collaborative team—this work reflects the efforts of researchers across disciplines and career stages, including three undergraduate students who are co-authors on this study!
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0
We are deeply grateful to all the patients and their families whose generosity made this study possible.
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0
This supports the conclusion that joint Hi-C/ONT-based reconstructions yield more accurate structural predictions than those based solely on short-read WGS.
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0

We compared ecDNA structure and transcription in two tumors: in P59, EGFR and CDK4 were predicted to be on separate ecDNAs; in P68, on the same ecDNA. Xenium data showed stronger EGFR-CDK4 expression correlation in P68 compared to p59.
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0


We found that ecDNA+ tumors form nuclear EGFR transcriptional hubs.
EGFR FISH on post-Xenium slides showed >70% of these hubs localized to the nucleus and overlapped with EGFR copy number - unlike control gene FABP7 with similar expression but no hub formation.
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0

We found that EGFR ecDNA tumors are hypoxic, mesenchymal-rich, and pericyte-enriched.
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0


Given our comprehensive whole-genome sequnecing datasets, we compared the spatial organization of ecDNA vs Linear Amplification GBMs
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0

We found that IDH-mutant gliomas frequently harbored inflammatory microglia expressing CX3CR1 specifically within AC-like malignant neighborhoods, rather than OPC-like niches—despite the OPC-like malignant cell state being more prevalent in IDH-mutant gliomas.
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0

To investigate the spatial tumor microenvironment of our glioma samples, we applied a single-cell spatial transcriptomics approach (10x Xenium), generating 4.6 million cells and over 775 million transcripts.
We identified distinct cell states in IDH-wildtype vs mutant gliomas
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0

We next asked whether treatment contributed to ecDNA loss in recurrences. Recurrent tumors arose from residual regions of the primary site, suggesting ecDNAs were spatially restricted subclones eliminated during initial surgery.
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0

We also examined EGFR expression levels, which showed a marked reduction in the recurrent tumors. Similarly, single-nucleus whole-genome analysis revealed the disappearance of the focal amplification on chromosome 7, while clonal alterations were retained.
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0

Hi-C and WGS data revealed the loss of ecDNA signals in recurrent tumor (P-12r1) while preserving other structural variations, such as chromothripsis on chr 3
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0

We also identified two GBM patients (P12 and P29) in whom ecDNA molecules were not implicated in tumor recurrence and were absent in both subsequent surgical specimens.
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0

Hi-C and long-read data were essential to improve ecDNA reconstructions.
For example, for p59, the short-read reconstruction suggested EGFR and CDK4 genes are on the same ecDNA circle.
However, Hi-C and ONT WGS-based reconstructions suggested these genes reside on two distinct circles.
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0


🔹 Resolving ecDNA Structure and Function
We used short- and long-read whole-genome sequencing plus Hi-C to reconstruct complex extrachromosomal DNA (ecDNA) structures. This revealed:
Tumors can harbor multiple ecDNA circles with distinct oncogenes.
22.05.2025 17:37 — 👍 0 🔁 0 💬 1 📌 0
We performed an integrative multi-omic and spatial transcriptomic analysis of 93 gliomas, leveraging 449 unique specimens to uncover how oncogenic drivers reshape tumor architecture and the surrounding microenvironment in human gliomas.
22.05.2025 17:37 — 👍 2 🔁 0 💬 1 📌 0

Excited to share our latest manuscript www.biorxiv.org/content/10.1...
This was a great collaboration with Drs. Huse, Lang, @jesserdixon.bsky.social
22.05.2025 17:37 — 👍 15 🔁 6 💬 1 📌 0
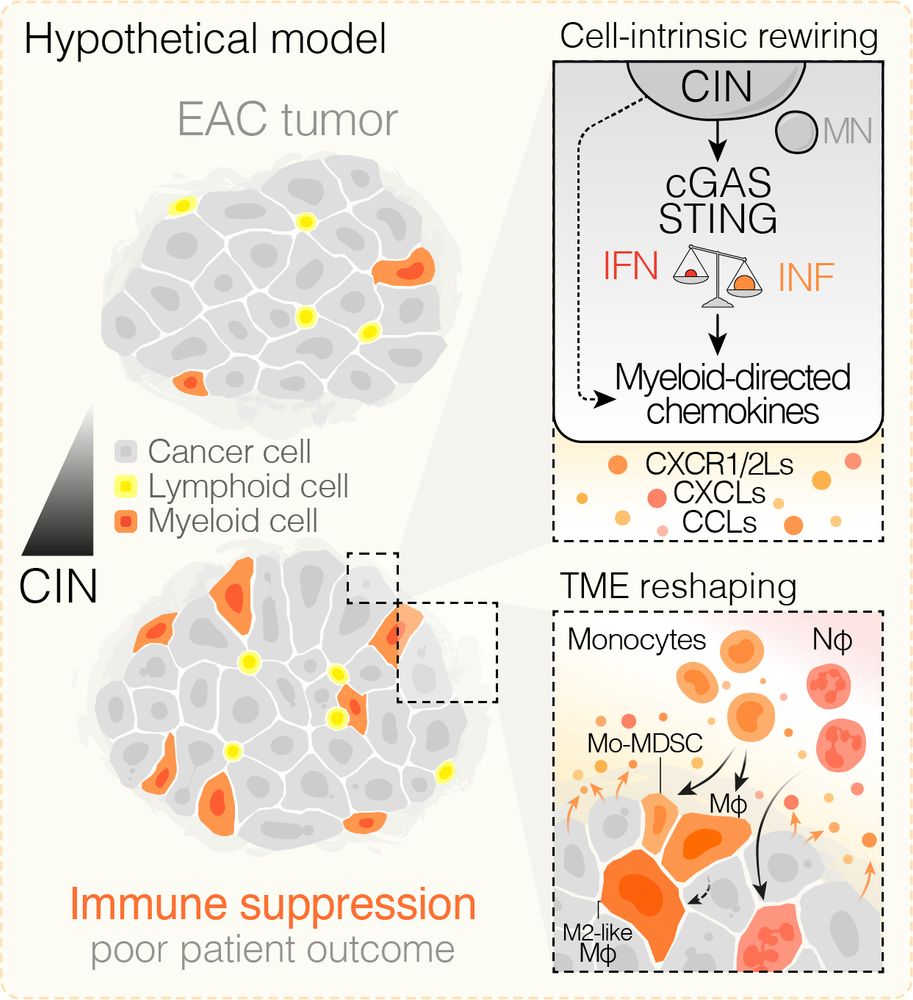
Model figure - in EAC cancers, as chromosomal instability (CIN) increases, we found that a CIN-driven cGAS-dependent axis drives myeloid directed chemokine expression and recruitment of tumor-favoring myeloid populations
Incoming - new preprint alert from the Parkes lab led by @brunobeernaert.bsky.social Full bluetutorial follows! 1/ 11 www.biorxiv.org/content/10.1...
12.05.2025 12:08 — 👍 47 🔁 19 💬 1 📌 2
Job Finder - Karriereportal - Universitätsklinikum Essen
Finden Sie eine Stelle am Universitätsklinikum Essen
🧪 We’re Hiring: Lab Technician 🧪
I’m happy to share that our newly established Emmy Noether Group in Essen is looking for a Lab Technician to join our team!
🔗 Apply here: karriere.ume.de/job-finder/?...
📅 Start: At the earliest possible date
💻 Homepage: kocakavuklab.vercel.app/overview
07.05.2025 20:32 — 👍 2 🔁 3 💬 0 📌 0
Omg, this is super funny but also very painful!!!
03.05.2025 18:56 — 👍 1 🔁 0 💬 0 📌 0
that’s right
26.04.2025 01:55 — 👍 59 🔁 11 💬 1 📌 0
Huge opportunity for postdocs gearing up for an academic job search, hosted by @stanford-chemh.bsky.social @berkeleymcb.bsky.social and UCSF!
25.04.2025 23:46 — 👍 51 🔁 34 💬 0 📌 0
YouTube video by Johan Gibcus
Rules of Engagement
Earnshaw, Goloborodko, Dekker & Mirny labs are excited to present our latest work, "Rules of engagement for condensins and cohesins guide mitotic chromosome formation" - now accepted!!
www.science.org/doi/10.1126/...
A short clip describing the key results:
www.youtube.com/watch?v=pmvO...
11.04.2025 07:45 — 👍 98 🔁 53 💬 7 📌 6
European Association of Neuro-oncology, multidisciplinary society, Clinical Trials, Research, Education #EANO2025 #btsm #neurooncology #braintumor #cancer | www.eano.eu
Mother to Sam Alp, Assistant professor @UTexasSPH Alumni: Izmir Science High School, Sabanci, Harvard, UTHealth, BCM
Studying cellular #stress responses, particularly #senescence and its impact on #immune response, #ageing and #cancer, #tumorigenesis at Cancer Research UK CI @cruk-ci.bsky.social, University of Cambridge @cam.ac.uk
Website: naritalab.com
Transcription factors, gene regulation, single cell, perturbations. Science, mountain and wilderness enthusiast. Assistant Professor @ University of Zurich
assistant professor @mskcancercenter.bsky.social
clareaulab.com
Group leader of Chromatin (Dys)function lab @MDC-BIMSB
Former Postdoc @Mundlos lab
PhD @ Schirmer lab
https://www.mdc-berlin.de/robson
Science Communicator and Visualization Specialist
✉ hello@joanaccarvalho.com
🔗 joanaccarvalho.com
I am the trick my mother played on the world.
Xennial Meteorologist in Houston. Forecasts: theeyewall.com & spacecityweather.com. Thoughts: mattlanza.substack.com
I like books, history, sports cards, baseball, Rutgers, #BHAFC.
Opinions my own.
Assistant professor @TGen | Postdoc @jacksonlab @MDAndersonNews | Genomics | Cancer | Gliomas | Telomeres
Principal Investigator
Lead of Cancer Metabolism Lab at @NCI.
Studying lipid metabolism in brain tumors.
Views are my own.
Developmental Biologist working at the Francis Crick Institute. Neural tube, morphogens, and gene regulatory networks. Editor-in-chief, Development.
London · briscoelab.org
Neuroscience Professor at UKBonn&DZNE Bonn. Bridging the gap between brain and immunity with genomics 🏳️🌈🏴☠️🇨🇳🇹🇷🇫🇷🇳🇪🇩🇪
ORCID 0000-0001-6319-404X
The Goodell Lab. Hematopoietic stem cells, epigenetics, DNA methylation. Houston Texas. Posts are my own.
Professor, Neurosurgeon Scientist
Mum of Twins, NIH and DOD funded
#WomenInStem #ASCI #AIMBE #EndCancer
Texas Ex - MD Anderson Alumni
The mission of the American Association for Cancer Research (AACR) is to prevent and cure cancer through research, education, communication, collaboration, science policy and advocacy, and funding for cancer research.






















