We committed ourselves to push new TCR-epitope data to our database every quarter.
Last week we published a new version of ImmuneWatch DETECT with TCRs for 75 different epitopes, of which 49 were not yet in our database.
Many new HPV epitopes!
immunewatch.gitlab.io/detect-docs/...
25.09.2025 11:47 —
👍 4
🔁 0
💬 0
📌 1
YouTube video by ImmuneWatch
Webinar: Annotate the specificity of your TCRs on Platforma
Analysing and annotating the specificity of TCR repertoires just became a lot easier.
Check out our joint webinar with @milaboratories.bsky.social on how to use ImmuneWatch DETECT within Platforma
m.youtube.com/watch?v=ECpA...
25.04.2025 12:01 —
👍 2
🔁 1
💬 0
📌 0

Webinar: Annotate the specificity of your TCRs on Platforma
Are you looking for a user-friendly way to analyse and annotate the specificity of your T-cell receptors?
Are you looking for a user-friendly way to analyse and annotate the specificity of your T-cell receptors?
Learn how you can use Platforma in combiination with ImmuneWatch DETECT in our webinar with @milaboratories.bsky.social CEO Stan Polavsky:
www.immunewatch.com/news/webinar...
31.03.2025 15:19 —
👍 3
🔁 0
💬 0
📌 0

Are you a bioinformatician working on T-cell Receptor (TCR) repertoires and are you looking for a state-of-the-art bioinformatics tool for antigen annotations? Then join us for our webinar:
Annotating the antigen specificity of a TCR repertoire.
xvx5k3zru3y.typeform.com/to/bvHafjzj?...
14.02.2025 11:07 —
👍 3
🔁 1
💬 0
📌 0
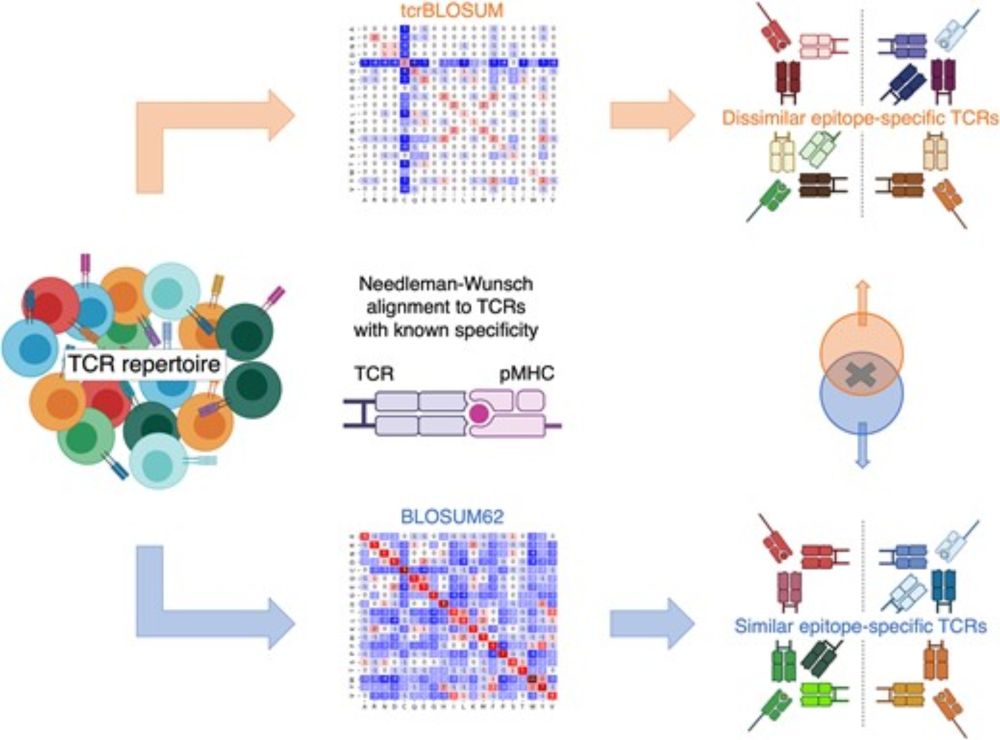
tcrBLOSUM: an amino acid substitution matrix for sensitive alignment of distant epitope-specific TCRs
Abstract. Deciphering the specificity of T-cell receptor (TCR) repertoires is crucial for monitoring adaptive immune responses and developing targeted immu
ImmuneWatch cofounders @pmeysman.bsky.social and @krislaukens.bsky.social have introduced tcrBLOSUM a specialised amino acid substitution matrix designed to enhance TCR repertoire analysis.
They found that tcrBLOSUM can improve the sensitivity of TCR clustering.
academic.oup.com/bib/article/...
14.01.2025 12:52 —
👍 4
🔁 0
💬 0
📌 0