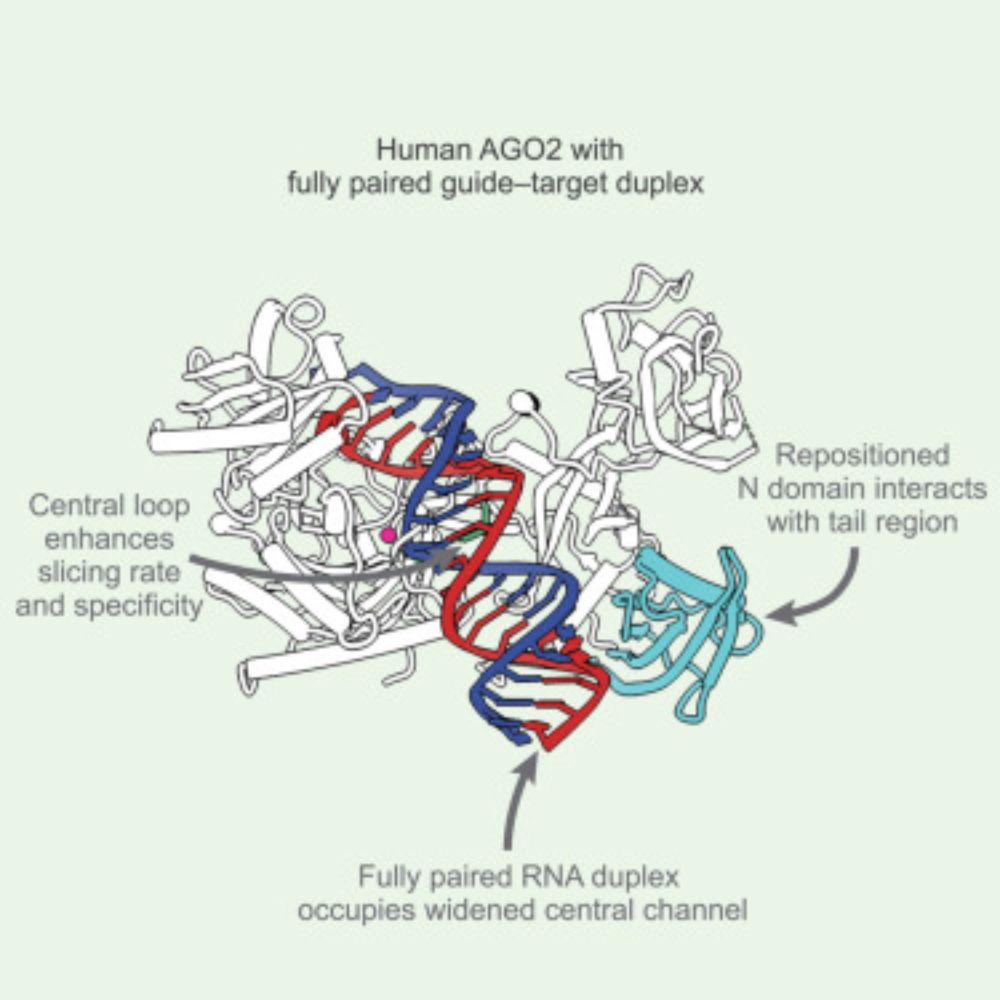
The GBM Compact - Focus on Condensate Biology poster session has 5 winners: Miriam Linsenmeier (FEBS Open Bio award), Patrick McCall and Michal Kacper Bialobrzewski (sponsored by De Gruyter Brill). Two book vouchers from Wiley were awarded to Saskia Hutten and Nora Haanaes! Congratulations!
22.09.2025 13:18 —
👍 7
🔁 3
💬 0
📌 2
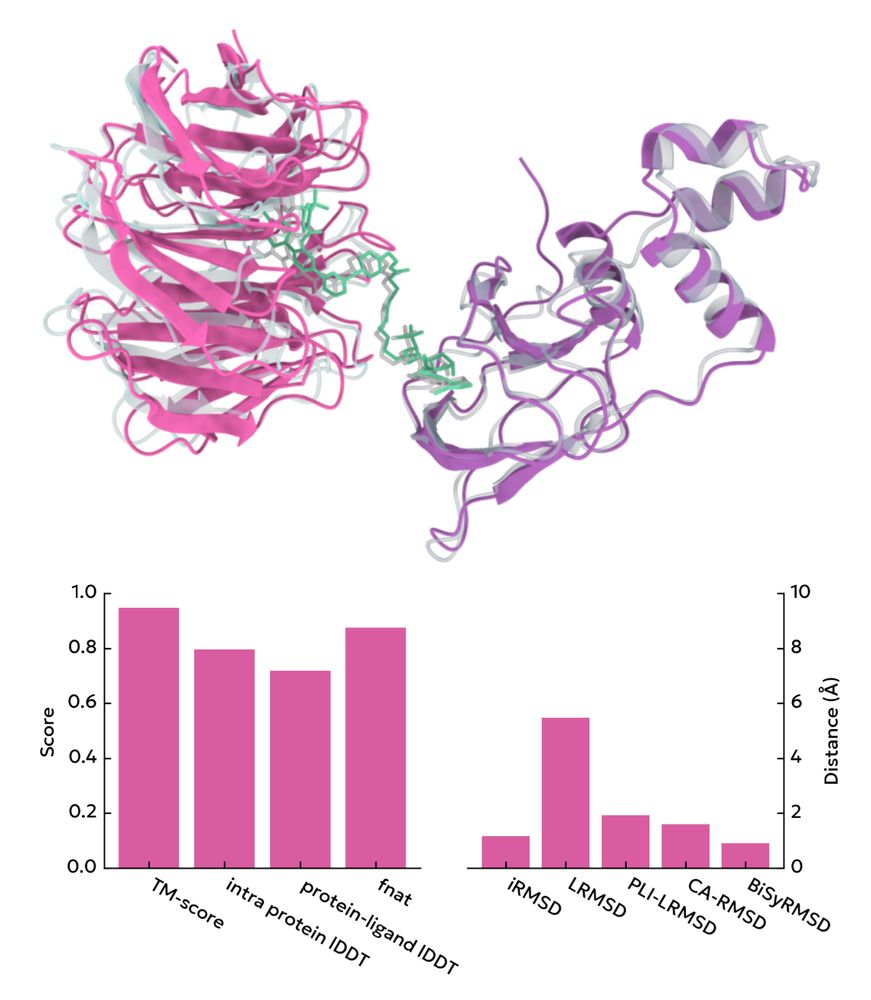
Screencap from the homepage showing various structural similarity metrics between a pose and a reference structure
A single python package for calculation of quality metrics like DockQ, LDDT, RMSD, & others
peppr.vant.ai
04.04.2025 06:32 —
👍 75
🔁 20
💬 3
📌 0
This is a massive public health and educational crisis, and we (scientists and journalist) need to help people make sense of it. Rapidly. Get your frames together people. Start sharing them in any way you can.
08.02.2025 01:36 —
👍 560
🔁 93
💬 9
📌 1


🚨 Revolutionising Snakebite Treatments with AI-Designed Proteins 🐍
I'm proud to share our latest study published in hashtag#Nature, driven by Susana Vazquez Torres, and co-led by David Baker (Institute for Protein Design, University of Washington) and myself.
15.01.2025 20:16 —
👍 27
🔁 11
💬 2
📌 2
Kartvelo,Kheli Khmals Ikar 💪
28.12.2024 21:01 —
👍 0
🔁 0
💬 0
📌 0
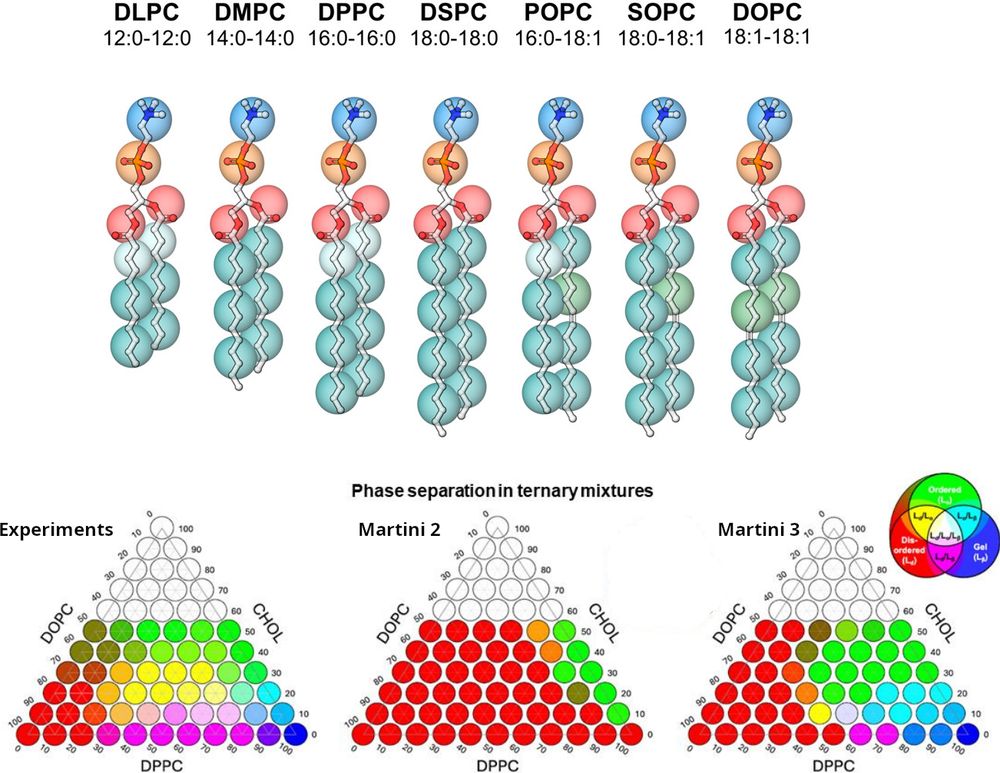
Excited to announce the release of our preprint, "The Martini 3 Lipidome: Expanded and Refined Parameters Improve Lipid Phase Behavior", now available on @chemrxiv.bsky.social
👉 Read the full preprint here: chemrxiv.org/engage/chemr...
26.12.2024 13:47 —
👍 56
🔁 14
💬 3
📌 2

Debunked dogma: disordered proteins disregard ligands’ chirality
Understanding unfolded proteins could boost drug discovery and decipher origins of life mysteries
Debunked dogma: disordered proteins disregard ligands’ chirality 🧬 my latest story for @chemistryworld.bsky.social on a fantastic and surprising Nature paper by Johan Olsen and Birthe Kragelund teams 🇩🇰 Read more below! #realtimechem #chemsky 🧪 www.chemistryworld.com/news/debunke...
18.12.2024 11:43 —
👍 80
🔁 13
💬 2
📌 2

AlphaFold3 still tends to overfit regions that AF2 multimer predicts to be disordered. Almost all disordered regions become helices in AF3.
15.12.2024 02:03 —
👍 55
🔁 20
💬 1
📌 2
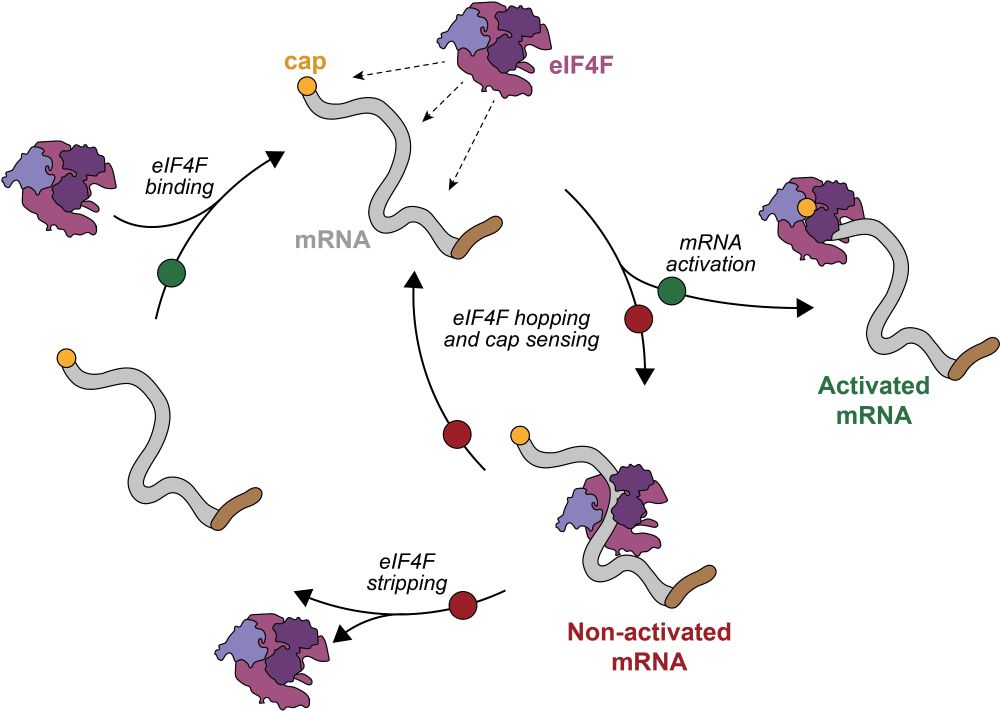
Our latest work is out in Nature today! Using smFRET, we directly visualized recruitment of the eIF4F complex to the 5' cap of eukaryotic mRNAs and formation of an activated mRNA. Our findings reveal new and surprising roles for each eIF4F component. 1/3 www.nature.com/articles/s41...
11.12.2024 21:35 —
👍 171
🔁 55
💬 11
📌 4
Congratulations @guyteichman.bsky.social et al!!
05.12.2024 22:22 —
👍 3
🔁 3
💬 0
📌 0