Here’s the updated Computational Biology Starter Pack! Let me know if you'd like to be included.
go.bsky.app/QVPoZXp
24.11.2024 16:33 — 👍 175 🔁 77 💬 81 📌 5
i would like to be added if possible!
25.11.2024 01:59 — 👍 1 🔁 0 💬 0 📌 0
New starter pack! go.bsky.app/GZ4hZzu
28.10.2024 09:43 — 👍 42 🔁 17 💬 6 📌 5
thanks!
21.11.2024 16:05 — 👍 0 🔁 0 💬 0 📌 0
could I be added? Thanks :)
21.11.2024 16:00 — 👍 1 🔁 0 💬 1 📌 0

Out-of-Distribution Validation for Bioactivity Prediction in Drug...
Recent advances in machine learning for materials science have significantly improved the prediction of novel materials. Building on these methods, we have adapted them for drug discovery...
We translated a validation strategy in Materials to Small molecules.
A key problem in cross-validation is splitting of data. Here we propose a step-forward CV. People are used to scaffold splits, time splits etc. but these all have limitations.
21.11.2024 02:04 — 👍 17 🔁 3 💬 1 📌 0
Two BioML starter packs now:
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
DM if you want to be included (or nominate people who should be!)
18.11.2024 17:09 — 👍 119 🔁 56 💬 10 📌 11
A starter pack for anyone interested in AI & drug discovery :)
go.bsky.app/AgYHc8j
15.11.2024 00:29 — 👍 53 🔁 15 💬 29 📌 2
thanks!
19.11.2024 13:43 — 👍 1 🔁 0 💬 0 📌 0
hi! i would like to be added if possible
17.11.2024 19:40 — 👍 0 🔁 0 💬 1 📌 0
thanks!
17.11.2024 18:53 — 👍 0 🔁 0 💬 0 📌 0
hi! could you add me :)
17.11.2024 18:51 — 👍 1 🔁 0 💬 1 📌 0
I'm making a list of AI for Science researchers on bluesky — let me know if I missed you / if you'd like to join!
go.bsky.app/AcP9Lix
10.11.2024 00:11 — 👍 246 🔁 90 💬 160 📌 5
This pack is now up to 67 people, broadly in AI and advanced data for science. Follow along, and forward me others who should be there! 🦋🦋
go.bsky.app/JeFdryY
16.11.2024 16:32 — 👍 45 🔁 15 💬 22 📌 0
We got an 🥂 Outstanding Paper Award!! Cannot be more grateful 🥹 This is super validating for our long pursuit of computational work on QUD.
Congrats to the amazing @yatingwu.bsky.social, Ritika Mangla, Alex Dimakis, @gregdnlp.bsky.social
15.11.2024 13:12 — 👍 60 🔁 9 💬 1 📌 0
thanks!
16.11.2024 04:20 — 👍 1 🔁 0 💬 0 📌 0
would like to be added :)
16.11.2024 04:08 — 👍 1 🔁 0 💬 1 📌 0
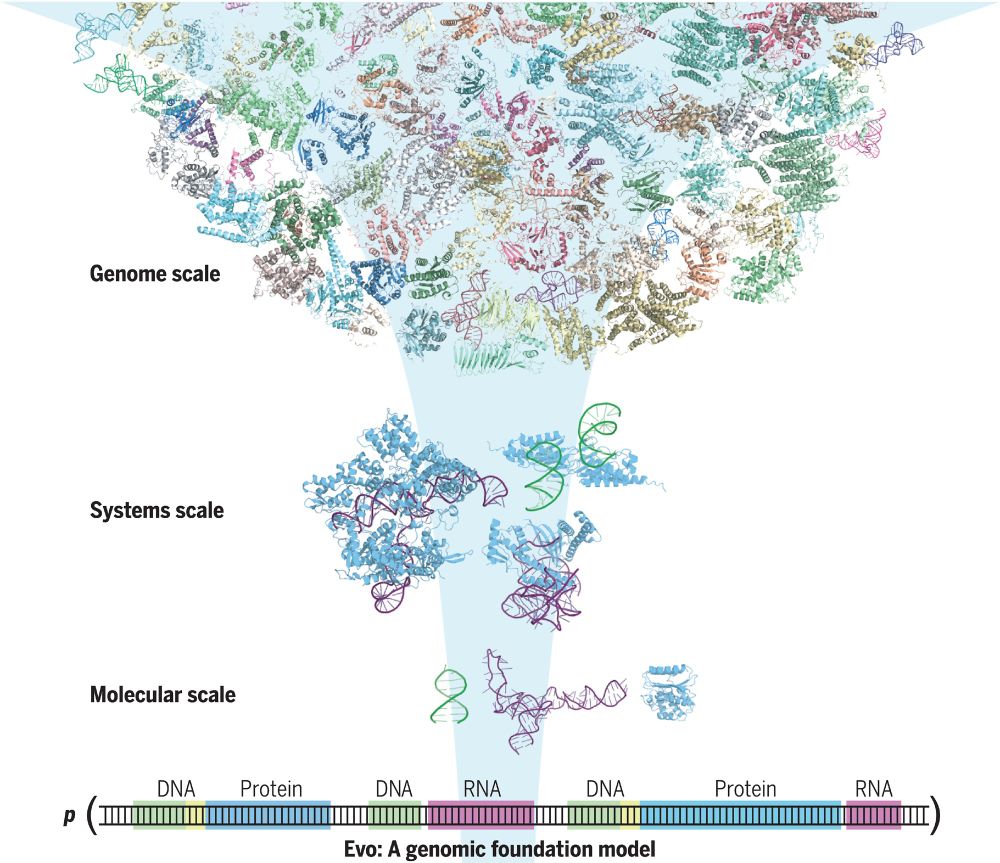
Evo, a 7-billion-parameter genomic foundation model, learns biological complexity from individual nucleotides to whole genomes.
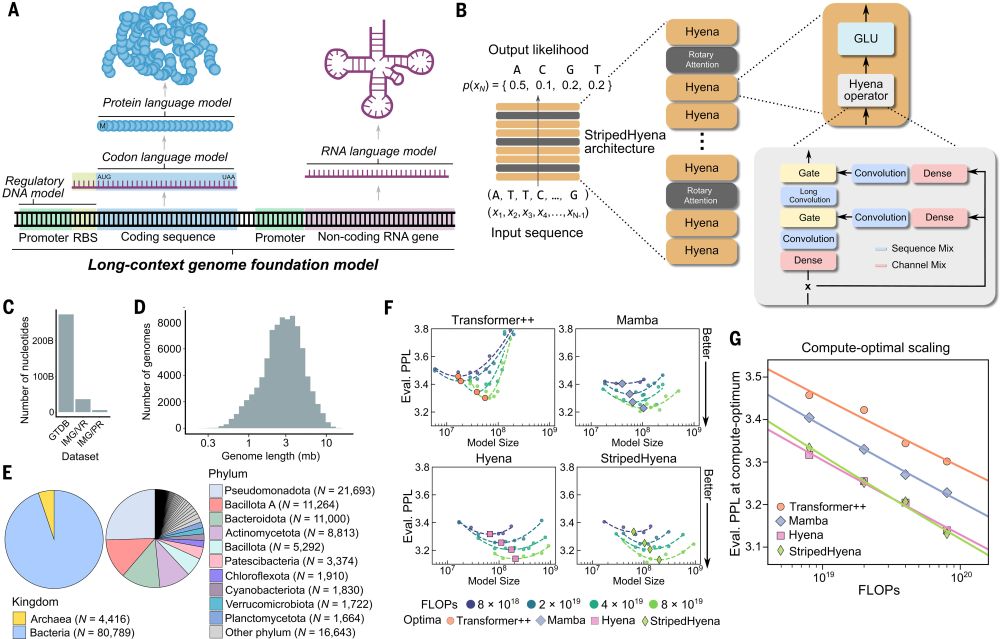
Pretraining a genomic foundation model across prokaryotic life.
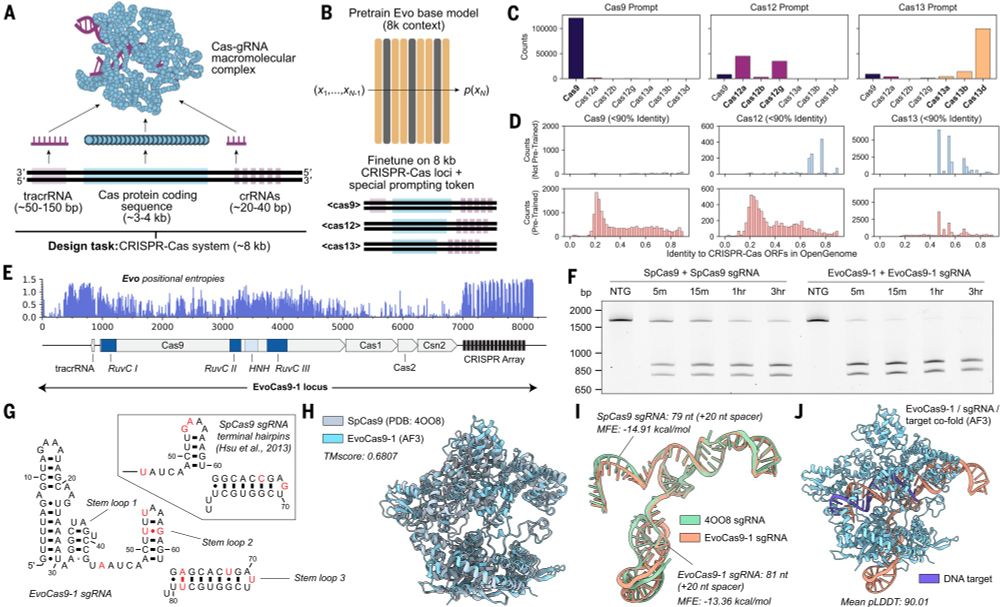
Fine-tuning on CRISPR-Cas sequences enables generative design of protein-RNA complexes.
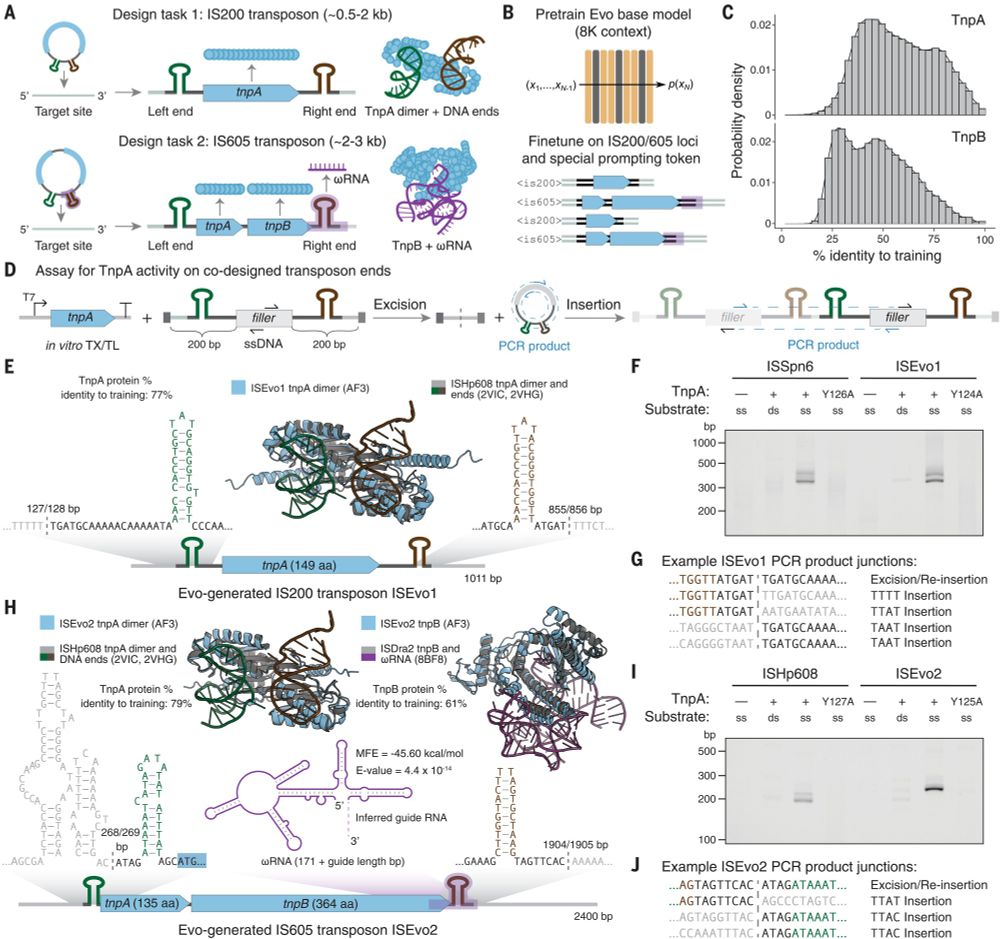
Fig. 4. Fine-tuning on IS200/IS605 sequences enables generative design of transposable biological systems.
Evo: A genomic language model of prokaryote genomes generates functional cas9 proteins and transposons.
@brianhiestand.bsky.social
www.science.org/doi/10.1126/...
14.11.2024 20:53 — 👍 59 🔁 27 💬 0 📌 1
would like to be on this list as well!
16.11.2024 00:50 — 👍 0 🔁 0 💬 1 📌 0
AI researcher at Google DeepMind -- views on here are my own.
Interested in cognition & AI, consciousness, ethics, figuring out the future.
🔬 Assistant Prof, Pathology @Duke | Director, Clin Micro Lab
🧫 Former Clin Micro Fellow @Memorial Sloan Kettering
👩🏻🔬 Former Postdoc @broadinstitute.org
🎓 PhD @The Rockefeller University
Focus: Diagnostics, AMR, Structural Biology
Senior Lecturer Microbial Genomics Newcastle University
Co-Founder and CTO Change Bio | PhD Engineering Biology Imperial College
Computational Biology-Chemistry-Medicine Postdoc at University of Zurich, she/they
Lifelong relationship with microbes, now studying them for a living. Assistant Professor at MedUni Graz. Opinions are my own. He/him 🦠+💻=❤️
More socials at cdiener.com. Lab website at dienerlab.com.
Deep learning, computational chemistry, generative modeling, AI for Science. Principal Research Manager at Microsoft Research AI for Science.
Senior Research Scientist at Google DeepMind. Prev @CarnegieMellon. IIT Indore Gold Medalist.
Principal Research Scientist at NVIDIA | Former Physicist | Deep Generative Learning | https://karstenkreis.github.io/
Opinions are my own.
Senior Research Director at Google DeepMind in our San Francisco office. I created Magenta (magenta.withgoogle.com) and sometimes find time to be a musician.
🏳️🌈Professor - Federal University of Uberlândia, Bioinorganic and (Bio)polymer Chemistry (views of my own).
(1) https://orcid.org/0000-0002-8357-3044
(2) https://www.linkedin.com/in/fernando-bergamini-7b577615/
(3) https://www.instagram.com/bioinspiredlab/
Head of the Centre for Artificial Intelligence, DS&AI, Biopharma R&D, AstraZeneca Views are my own
Assistant Professor at UT Southwestern Medical Center | Full-time reproductive biologist | Part-time genomics technology developer | Lab website: https://haiqichenlab.github.io/
Assoc Prof @UWaterloo
Research:
#ArtificialIntelligence #MachineLearning #ReinforcementLearning
#casual-learning, dimensionality reduction, #ethicalAI #aimorality #aialignment
Domains: wildfires, driving, medical, lidar
Diversity is Strength.
AI at Google DeepMind. Previously: Orbital Insight, A9/Amazon.
adamwkraft.github.io
Research Scientist GoogleDeepMind
Ex @UniofOxford, AIatMeta, GoogleAI
ML. Biology. Research Engineer at Google DeepMind.
Google Chief Scientist, Gemini Lead. Opinions stated here are my own, not those of Google. Gemini, TensorFlow, MapReduce, Bigtable, Spanner, ML things, ...
Research Scientist at Google DeepMind
Gemini evals and post-training
sebarnold.net





