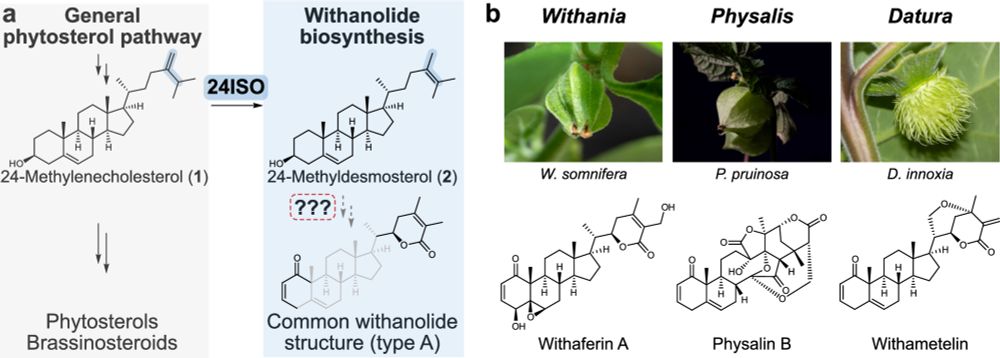
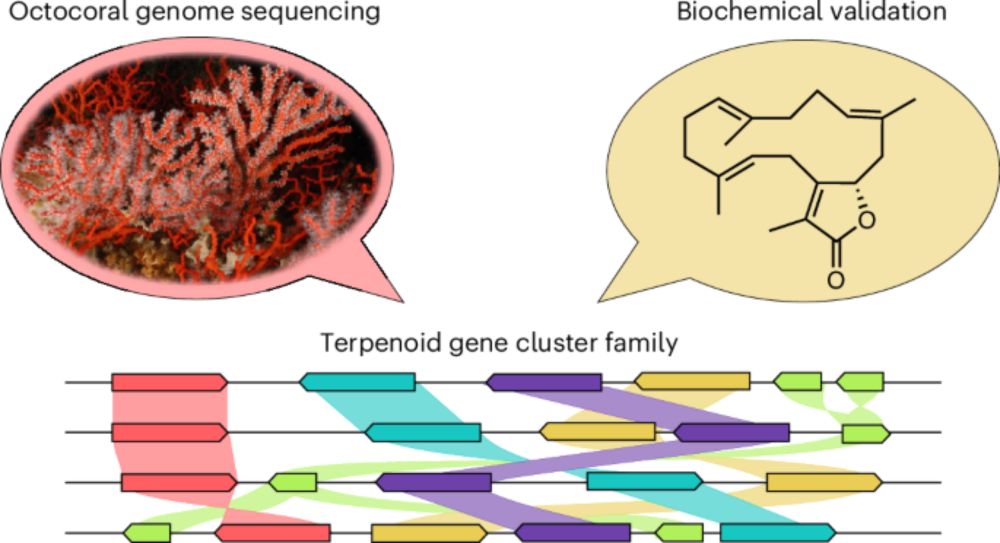
Chemical richness and diversity of uncultivated ‘Entotheonella’ symbionts in marine sponges - Nature Chemical Biology
Marine sponges host bacteria that produce diverse bioactive compounds. Here, the authors conduct a large-scale metagenomic, single-bacterial and biochemical study to reveal the untapped biosynthetic p...
Happy to share work by Maria and awesome collaborators published in Nature Chem Bio!
Using metagenomic, single-bacterial, and biochemical methods we provide insights into uncultivated bacterial symbionts as rich sources of bioactive natural products in marine sponges.
#DrugDiscovery #NaturalProducts
18.11.2025 11:44 — 👍 5 🔁 1 💬 0 📌 0

Discovery of Late Intermediates in Methylenomycin Biosynthesis Active against Drug-Resistant Gram-Positive Bacterial Pathogens
The methylenomycins are highly functionalized cyclopentanone antibiotics produced by Streptomyces coelicolor A3(2). A biosynthetic pathway to the methylenomycins has been proposed based on sequence analysis of the proteins encoded by the methylenomycin biosynthetic gene cluster and the incorporation of labeled precursors. However, the roles played by putative biosynthetic enzymes remain experimentally uninvestigated. Here, the biosynthetic functions of enzymes encoded by mmyD, mmyO, mmyF, and mmyE were investigated by creating in-frame deletions in each gene and investigating the effect on methylenomycin production. No methylenomycin-related metabolites were produced by the mmyD mutant, consistent with the proposed role of MmyD in an early biosynthetic step. The production of methylenomycin A, but not methylenomycin C, was abolished in the mmyF and mmyO mutants, consistent with the corresponding enzymes catalyzing the epoxidation of methylenomycin C, as previously proposed. Expression of mmyF and mmyO in a S. coelicolor M145 derivative engineered to express mmr, which confers methylenomycin resistance, enabled the resulting strain to convert methylenomycin C to methylenomycin A, confirming this hypothesis. A novel metabolite (premethylenomycin C), which readily cyclizes to form the corresponding butanolide (premethylenomycin C lactone), accumulated in the mmyE mutant, indicating the corresponding enzyme is involved in introducing the exomethylene group into methylenomycin C. Remarkably, both premethylenomycin C and its lactone precursor were one to two orders of magnitude more active against various Gram-positive bacteria, including antibiotic-resistant Staphylococcus aureus and Enterococcus faecium isolates, than methylenomycins A and C, providing a promising starting point for the development of novel antibiotics to combat antimicrobial resistance.
So cool. Greg Challis shows that a methylenomycin precursor is a better antibiotic than methylenomycin itself. This is in S.coelicolor A3(2)! Probably many similar examples yet to be discovered of pathways intermediates being useful, but consistently overlooked. #secmet
pubs.acs.org/doi/10.1021/...
04.11.2025 18:38 — 👍 11 🔁 2 💬 0 📌 0

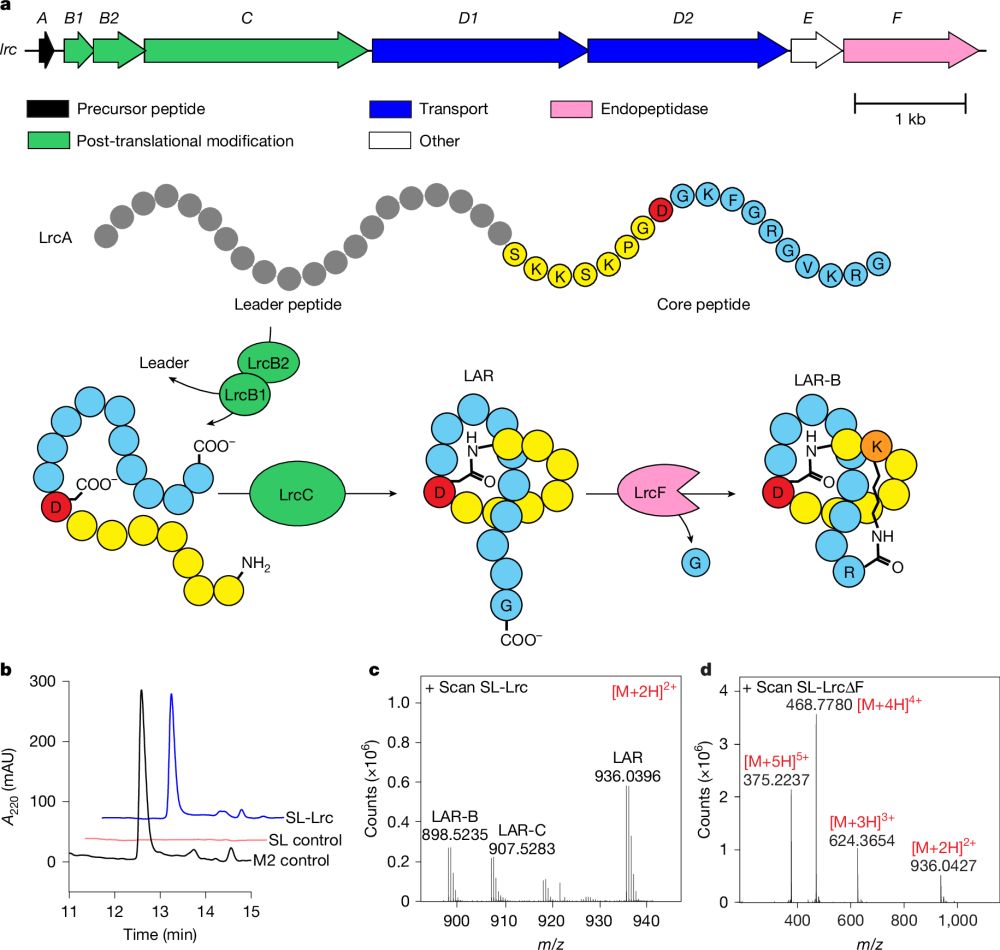
Growth-coupled microbial biosynthesis of the animal pigment xanthommatin — Led by Leah Bushin, featuring M. Gracia Alvan, @danielvolke.bsky.social @oscarpuiggene.bsky.social in a fantastic collaboration w/Brad Moore @labnikel.bsky.social @natbiotech.nature.com 🦑
www.nature.com/articles/s41...
03.11.2025 12:49 — 👍 33 🔁 15 💬 2 📌 1
University of Florida - Details - Assistant/Associate Professor in Marine Bioscience
Marine Biosciences Assistant/Associate Prof job at University of Florida explore.jobs.ufl.edu/en-us/job/53... [for MNP folks, this is where Sansa Loesgen @loesgenlab.bsky.social is based]
27.10.2025 15:10 — 👍 4 🔁 3 💬 0 📌 0
IDBac: an open-access web platform and compendium for the identification of bacteria by MALDI-TOF mass spectrometry. https://www.biorxiv.org/content/10.1101/2025.10.15.682631v1
16.10.2025 02:17 — 👍 3 🔁 3 💬 0 📌 0

Trends in metabolite discovery from Actinomycetes
Covering: 2013 to 2023 In this review, we analyzed the scientific literature of the period 2013–2023 that reported novel specialized metabolites from the Actinomycetes, one of the most prolific…
Be sure to read this review, part of our Industrial Perspective themed collection, by Stefano Donadio & co. from NAICONS Srl discussing the trends in metabolite discovery from Actinomycetes #secmet #natprod
Find it in full below👇
26.09.2025 11:56 — 👍 13 🔁 7 💬 0 📌 0
Awesome work, @mmzdouc.bsky.social !
27.09.2025 09:13 — 👍 6 🔁 1 💬 0 📌 0
(I have one too! Probably lots of overlap...)
And check out the Secondary Metabolism feed. Lots of awesome science here on Bluesky!
bsky.app/profile/did:...
24.09.2025 22:52 — 👍 2 🔁 1 💬 0 📌 0

@mitjare.bsky.social kicking off the second day of the VAAM Workshop at @freieuniversitaet.bsky.social talking about the phyllosphere, its bacterial inhabitants and the natural products they produce - including surfactants!
25.09.2025 06:42 — 👍 5 🔁 1 💬 1 📌 0
Congrats to the poster prize!🥳
27.09.2025 19:25 — 👍 1 🔁 0 💬 0 📌 0

Just wrapped up inspiring days of science and networking at the VAAM Workshop “Biology of Bacteria Producing Natural Products” in Berlin!
Our team’s first conference ended on a high note — 2nd prize for best poster! 🥈🥳
Thanks to the organizers @niedermeyer-lab.bsky.social for a fantastic meeting. 🙏
26.09.2025 13:09 — 👍 12 🔁 3 💬 2 📌 0
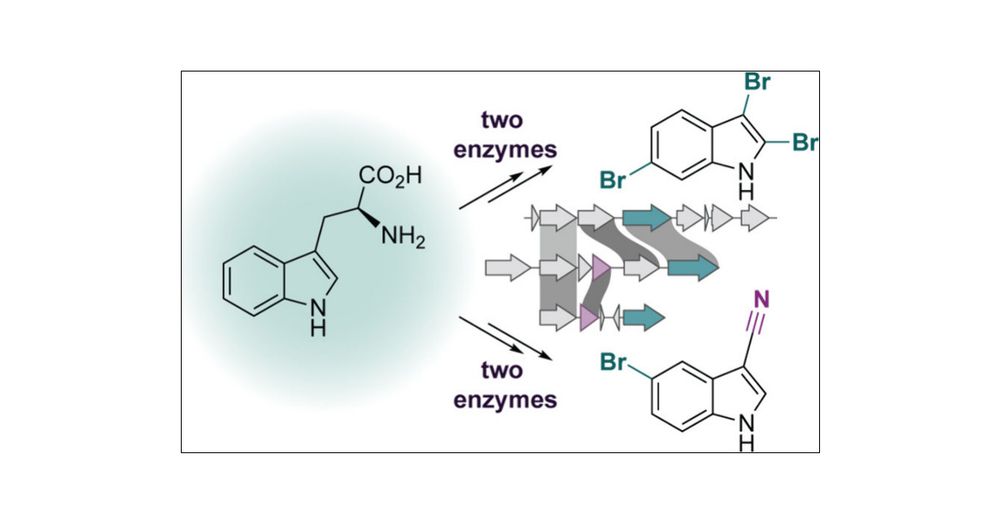
Metagenomic Identification of Brominated Indole Biosynthetic Machinery from Cyanobacteria
Halogenated indole natural products have been isolated from a variety of organisms, including plants, marine algae, marine invertebrates, and bacteria. Aquatic cyanobacteria, in particular, are rich producers of brominated indoles, but their cognate biosynthetic enzymes have only been successfully linked in a limited number of natural products, such as the eagle-killing toxin aetokthonotoxin (AETX). The biosynthetic pathway for AETX involves five enzymes, two of which were previously undescribed due to incomplete annotations as hypothetical proteins. Our recent elucidation of AETX biosynthesis established functions of the two previously unknown proteins as enzymes responsible for tryptophan halogenation (AetF) and nitrile synthesis (AetD). Given their sequence novelty, we queried metagenomic data sets for these two enzymes and identified two new cyanobacterial haloindole biosynthetic gene clusters (BGCs) from marine sediment in Moorea, French Polynesia, and soil-derived samples in Maunawili Falls, Hawaii. We characterized the recovered BGCs by biochemically validating a new AetF homologue that exclusively halogenates free indole, rather than tryptophan as observed in AETX biosynthesis, and a new AetD homologue that harbors distinct substrate preferences, expanding the scope of nitrile biosynthesis. Additional characterization of core and accessory enzymes within these AETX-like BGCs highlights the breadth and diversity of haloindole biosynthetic machinery in cyanobacteria.
New work led by the amazing @aprillukowski.bsky.social lab. Love working on these interdisciplinary projects discovering new chemical diversity in nature. Microbes are the best chemists!
pubs.acs.org/doi/10.1021/...
12.07.2025 16:30 — 👍 7 🔁 3 💬 0 📌 0

On the left, a picture of a Petri dish showing an amycalotopsis from which comes in the center, a cartoon dragon and under the dragon the molecules called Dracomycins, which are drawn in the shape of the dragon. On the right a petri dish showing a cartoon of zones of inhibition.
Dracomicins, Hybrid Oligosaccharide–Nonribosomal Peptide Antibiotics from Amycolatopsis Species
pubs.acs.org/doi/10.1021/...
13.06.2025 21:40 — 👍 11 🔁 4 💬 0 📌 1

Save the date!
13.06.2025 15:33 — 👍 18 🔁 4 💬 1 📌 1
I am thrilled to share after years of work/procrastination that the MassQL manuscript is finally published in @natmethods.nature.com - "A universal language for finding mass spectrometry data patterns". This was an team effort from all co-authors that helped shape MassQL and how it could be used.
12.05.2025 18:10 — 👍 40 🔁 17 💬 1 📌 2
It’s not every day one gets to publish an article with NASA astronauts. This journey started in 2019 and withe the goal to understand the microbial and chemical make-up of the space station. We had lots of experience with analysis from swabs on earth.
27.02.2025 18:52 — 👍 42 🔁 17 💬 1 📌 1
BIOspektrum - Das Magazin für Biowissenschaften * Neue Themen aus der Wissenschaft * Nachrichten aus der Scientific Community
http://www.biospektrum.de
Junior Research Group "Metabolomics-guided Natural Product Discovery" @leibniz-hki.de, Jena, Germany.
Head: @jethrohemmann.bsky.social
https://www.leibniz-hki.de/en/metabolomics-guided-natural-product-discover.html
Assistant Professor of Medicinal Chemistry at ETH Zürich
Scientist and all around amateur.
professor @ Rose-Hulman Institute of Technology👩🏫 • passionate about undergraduate research🧬 • biochemist👩🔬 • natural product biosynthesis⚗️ • wanderluster✈️ • triathlete🏊🏻♀️🚴🏻♀️🏃♀️ • cat person 🐈⬛
Bioinorganic #Chemsky Group of Lena Daumann @HHU.de PI tweeting. We love iron, rare earth elements, enzymes and bacteria!🧫 🧪 #fblockrocks www.bioac.hhu.de/
High impact, critical reviews in natural products research and related areas from RSC Publishing.
Published by @rsc.org 🌐 Website: rsc.li/natprodreports
Independent and non profit #Research Organization developing innovative programs, collaborations and contracts with academic and industrial partners worldwide.
Granada, Spain.
medinadiscovery.com
Interested in microbial ecology & evolution. Views are only my own. (he/him)
🎓: https://scholar.google.com/citations?hl=en&user=OBPpZq4AAAAJ&view_op=list_works&sortby=pubdate
👨💻: https://github.com/raufs
Natural Products Magnetic Resonance Database
https://np-mrd.org
Database and repository for natural products NMR data
We drive innovation in the discovery and bioanalytical characterization of natural products, leveraging state-of-the-art mass spectrometry, NMR spectroscopy and advanced bioinformatics, powered by AI-driven strategies.
Teaching molecular and cell biology, genetics and immunology at the Frankfurt Goethe University, Department Biochemisty, Chemistry and Pharmacy
Scientific topics are: diagnosis and cancer mechanisms in Acute Leukemia; contributed to SARS-CoV-2 research
Plants, specialized metabolism, synthetic biology, protein evolution, and everything in between.
Postdoc at Sattley & Fordyce Labs, Stanford
Previously SynBio @ Voigt & Laub Labs, MIT
Cryo-EM ❄️🔬 | Molecular machines & mechanisms 🧬 | Associate professor @oxfordbiochemistry ghilarovlab.com
PhD student in Biomolecular Chemistry @leibniz-hki.de. Interested in specialized metabolites involved in virulence of human-pathogenic bacteria.
PhD in Natural Products Chemistry. Now investigating cyanobacteria 🦠