
PanGene-O-Meter: Intra-Species Diversity Based on Gene-Content
Bacterial genome evolution is shaped to a great extent by horizontal gene transfer, detectable as genes with a presence-absence pattern of variation that does not follow phylogenetic relationships acr...
How diverse are the individual strains within your species?
How broad is their Pangenome?
How will you choose representatives from the whole collection?
Look no further! PanGene-O-Meter is here for you!
From @haimashkenazy.bsky.social @plantevolution.bsky.social
www.biorxiv.org/content/10.1...
02.09.2025 07:20 — 👍 31 🔁 19 💬 1 📌 0
First a pentamer, then a hexamer… now an octamer!
The riddle of the enigmatic CCG10-NLR immune receptor family cracked open 🔥
Congrats Guanghao @GuanghaoGuo He Zhang @mhz1989 Selva @M__Selvaraj et al.
#NLRbiology #plantsci #immunology
28.08.2025 12:05 — 👍 44 🔁 25 💬 1 📌 1
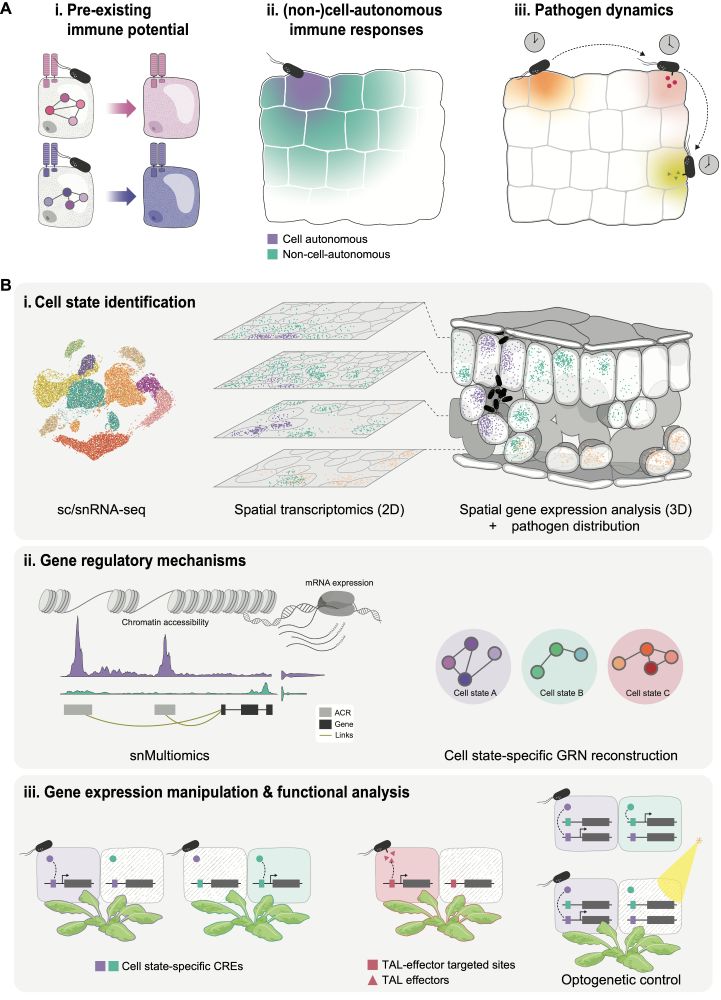
How can we better understand the plant immune system? In my new @cp-cellhostmicrobe.bsky.social piece, I propose “immune cell states” as a framework for uniting molecular and cellular perspectives.
www.cell.com/cell-host-mi...
13.08.2025 15:12 — 👍 80 🔁 39 💬 3 📌 3
Thank you Baptiste
04.08.2025 15:23 — 👍 0 🔁 0 💬 0 📌 0
This helps disentangle the complex relationship between plant development and immunity.
04.08.2025 07:44 — 👍 7 🔁 1 💬 0 📌 0

2/3 By focusing on very early time points, Shanshan found mesophyll cell populations that not only prioritize a defense-over-growth strategy vs. a growth-over-defense strategy, but also that these cell populations behave more similarly during ETI than PTI.
02.08.2025 14:50 — 👍 8 🔁 2 💬 1 📌 0
Super excited to share our new preprint! 🎉 This project has been an incredible learning experience, thanks to the amazing lab and wonderful people I get to work with. Huge thanks to my supervisor! @plantevolution.bsky.social
04.08.2025 07:41 — 👍 26 🔁 9 💬 1 📌 0
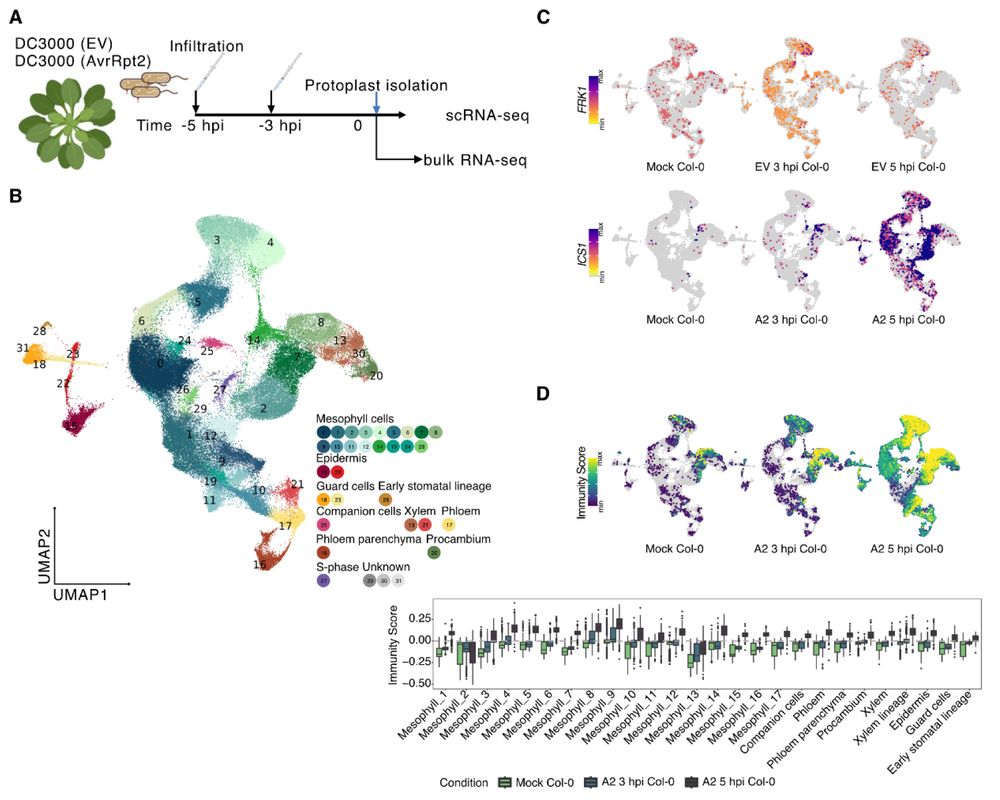
1/3 New preprint led by Shanshan Wang, in collaboration with the Timmermans lab.
Several groups have presented sc/snRNA-seq analyses of Arabidopsis leaves after bacterial infection before. Is there anything left to discover? Turns out: Yes.
#plantscience
www.biorxiv.org/content/10.1...
02.08.2025 14:46 — 👍 73 🔁 39 💬 2 📌 1
He/Him | GitHub-> https://github.com/omicscode | RUST 🦀Bioinformatician, Software Developer, AI/ML/DL, Data Engineer -> https://www.icloud.com/iclouddrive/088QrZHmTgS5_iJv8RpT1-yLg#GauravSablok | Read all Post & Replies | I don’t vibe code | Ignore typos.
PhDone👩🎓 of Schaller Lab at Uni-Hohenheim; 🍅 Systemin🐛;Plant immunity; Phosphoproteomics; R coding; She/her; 🐶🐱lover somehow adopt a 🦜;🍺 food-motivated; incoming postdoc
C4 grasses, hydraulics, single cell, plant physiology, anatomy and everything in between
📍 Gregory lab & Helliker lab, University of Pennsylvania.
Post doc in the Ecker lab @ The Salk Institute. PhD in the Bailey-Serres lab @ UCR. Posts include papers I'm reading and things that I 3D print
@TrALEE_Sci on Twitter
Plant diseases threaten ecosystems worldwide.
Our mission is to decode disease resistance mechanisms—a vital step toward safeguarding plant health and ensuring a sustainable future.
☞ A plant immunity lab focusing on gene regulation
☞ www.thedinglab.com
Biologist @ The Sainsbury Lab; passionate about plant pathogens & evolution; open science advocate; loves travel, food and sports; nomad and hunter-gatherer. Web http://kamounlab.net | Medium https://medium.com/@kamounlab | GetGenome http://getgenome.net
Postdoc in the @geminiteamlab.bsky.social ZMBP, Uni Tübingen
Plant - Microbe Interactions 🌱🦠🍄
She/her
Imperial College London. Exploring plant-microbe interactions & receptor-dependent immunity. Bioinformatics, evolution, synthetic biology. #PlantScience #Bioengineering
https://profiles.imperial.ac.uk/j.kourelis
Professor for Plant Cell Biology @ruhr-uni-bochum.de | #theustunlab | DFG Emmy Noether alumni | ERC StG #DIVERSIPHAGY | in 💚 with #proteostasis 🇺🇸🇪🇸🇫🇷🇩🇪🇹🇷🇮🇹🇨🇳🏳️🌈 He/Him
http://theustunlab.com/
@theustunlab.bsky.social
Or just “Li” |
Assist. Prof. @ Plant Bio Michigan State U. |
Also post data visualization |
Lab: https://cxli233.github.io/cxLi_lab/ |
GitHub: https://github.com/cxli233
Plants, specialized metabolism, synthetic biology, protein evolution, and everything in between.
Postdoc at Sattley & Fordyce Labs, Stanford
Previously SynBio @ Voigt & Laub Labs, MIT
Postdoc at @UBuffalo | Gokcumen Lab
Evolution, structural variants and population genetics.
PhD from @UABBarcelona | Inversion polymorphism.
https://biolevol.github.io/
Plant scientist and entrepreneur • Former postdoc @plantevolution.bsky.social • Now building Biogenda (http://biogenda.de) and The Custom-Lab Institute (https://custom-lab.de / @custom-lab.bsky.social)
Interested in fungi and plant-pathogen interactions🌱 currently working with @talbotlabtsl.bsky.social 🇮🇨🇦🇷🇬🇧
Postdoc in WeigelWorld @PlantEvolution, @MPI_bio. Evolution of Plant-Microbe-Microbe interactions, molecular evolution, and comparative genomics
Associate Professor | UNC Chapel Hill, Pharmacology Department | Lineberger Comprehensive Cancer Center | Research in Endocrinology, Epigenetics, & Wnt signaling| Views my own. https://www.med.unc.edu/pharm/pruittlab/team/
🇨🇳Professor@HZAU←🇩🇪GL@MPIPZ←🇺🇸PD@UofMinnesota←🇯🇵PhD/MS/BS@HokkaidoU
Plant-microbe interactions
Highly Cited Researcher (ISI Web of Science)
Chinese Government Friendship Award and Chime Bell Award
https://plantimmunity.hzau.edu.cn/
Star Trek, football lover
Departmental Group Leader - Plant Immunogenomics 🌱🧬🦠
Max Planck Institute for Biology - Tübingen 🇩🇪. Plant immunity and other small things




