Many people hours, calls and messages later: OSTA is now “in (pre)print”, though the real thing lives at bioconductor.org/books/OSTA.
Check it out, get in touch. We welcome any feedback, suggestions, wishes (& contributions).
It’s been a joy working with you @estellayixingdong.bsky.social!
21.11.2025 17:31 —
👍 40
🔁 24
💬 1
📌 4
Orchestrating Spatial Transcriptomics Analysis with Bioconductor https://www.biorxiv.org/content/10.1101/2025.11.20.688607v1
21.11.2025 14:46 —
👍 13
🔁 10
💬 0
📌 1
Huge thanks to @helucro.bsky.social and @markrobinsonca.bsky.social for great collaboration and supervision, and to the Robinsonlab and the @bioconductor.bsky.social community for valuable feedback throughout this project!
Feedback on the manuscript and package is welcome and much appreciated!
14.10.2025 15:15 —
👍 1
🔁 0
💬 0
📌 0
We highlight structure-based analysis using two publicly available datasets:
1. Quantifying structural rearrangements during colorectal malignancy transformation.
2. Recovery of structurally relevant gene expression gradients in human tonsil germinal centres.
14.10.2025 15:14 —
👍 0
🔁 0
💬 1
📌 0
Using our package sosta, we show how to reconstruct anatomical structures, quantify geometric features and other structurally-relevant characteristics, and compare features across samples and conditions.
14.10.2025 15:14 —
👍 0
🔁 0
💬 1
📌 0
Most spatial omics methods focus on single cells, but biological function often emerges from organised multicellular structures (like glands, crypts, or germinal centres)
14.10.2025 15:12 —
👍 0
🔁 0
💬 1
📌 0
I'm very excited to share our latest preprint!
We introduce structure-based analysis of spatial omics data – an approach that focuses on multi-cellular anatomical structures rather than single cells.
We also present sosta to facilitate this type of analysis: bioconductor.org/packages/sos...
14.10.2025 15:11 —
👍 6
🔁 3
💬 1
📌 0
Many thanks to everyone involved 🤝 Martin Emons, @helucro.bsky.social , Izaskun Mallona, Reinhard Furrer, @markrobinsonca.bsky.social and all Robinsonlab members.
04.12.2024 12:18 —
👍 0
🔁 0
💬 1
📌 0
Redirect to 00-home.html
We give an overview of established methods for the analysis of both lattice- and point pattern-based data and discuss common challenges. More information can be found in the accompanying website: robinsonlabuzh.github.io/pasta/
04.12.2024 12:17 —
👍 0
🔁 0
💬 1
📌 0
In lattice-based analysis we assume that the locations were fixed at the time of sampling and study the associated features at each location accounting for spatial relationships. This offers an observation-based on the data.
04.12.2024 12:16 —
👍 0
🔁 0
💬 1
📌 0
Point pattern-based analysis offers an event-based view of the data. It allows us to study the processes that lead to an pattern that is e.g., clustered.
04.12.2024 12:16 —
👍 0
🔁 0
💬 1
📌 0
This offers two streams of analysis: point pattern- or lattice-based analysis.
04.12.2024 12:16 —
👍 0
🔁 0
💬 1
📌 0
Imaging-based data can be viewed as a point pattern either in terms of transcript locations or cell centroids. Alternatively, the segmented cell outlines can be interpreted as an irregular lattice. HTS-based approaches are most often recorded on a regular lattice.
04.12.2024 12:16 —
👍 0
🔁 0
💬 1
📌 0
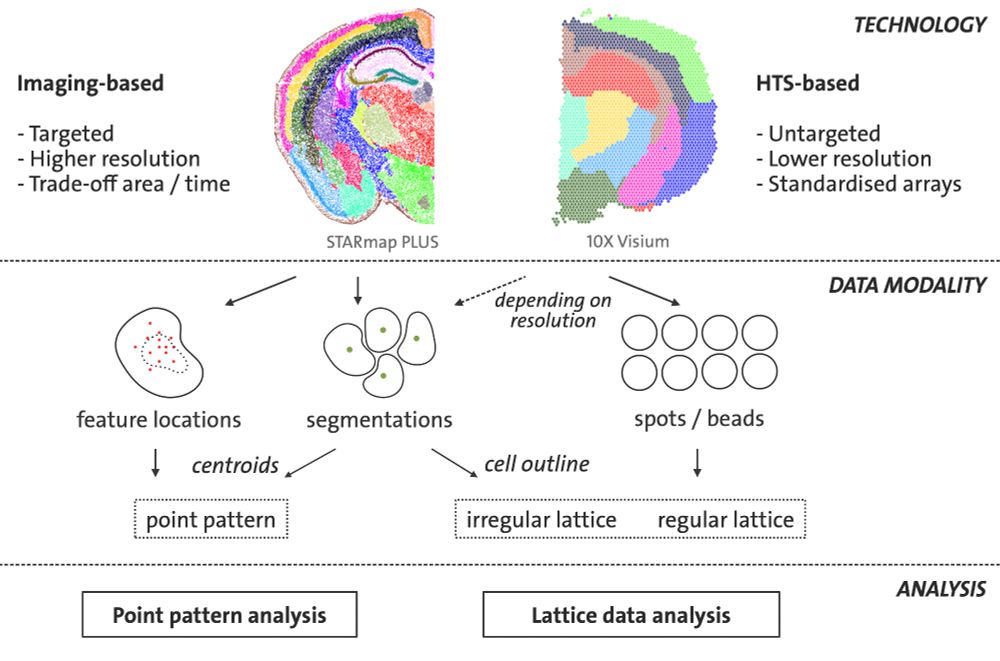
Spatial omics data can be classified into imaging-based and high throughput sequencing (HTS)-methods that differ in resolution and the number of features targeted.
04.12.2024 12:15 —
👍 0
🔁 0
💬 1
📌 0

pasta: Pattern Analysis for Spatial Omics Data
Spatial omics assays allow for the molecular characterisation of cells in their spatial context. Notably, the two main technological streams, imaging-based and high-throughput sequencing-based, can gi...
Interested in spatial statistics for spatial omics data? Check out our new resource: pasta.
We show how different technologies lead to different data modalities give and overview of point-pattern and lattice-based spatial analysis. Feedback welcome!
arxiv.org/abs/2412.01561
04.12.2024 12:14 —
👍 18
🔁 7
💬 1
📌 1




