⏱️ Ten days left. Time flies, so apply now 👉 www.protaiomics.eu
Don’t miss your chance to join ProtAIomics and a vibrant international community in AI powered proteomics.
#ResearchCareers #Proteomics #ArtificialIntelligence #MSCA #PhDOpportunities #FullyFundedPhD
21.11.2025 13:39 — 👍 4 🔁 2 💬 0 📌 0

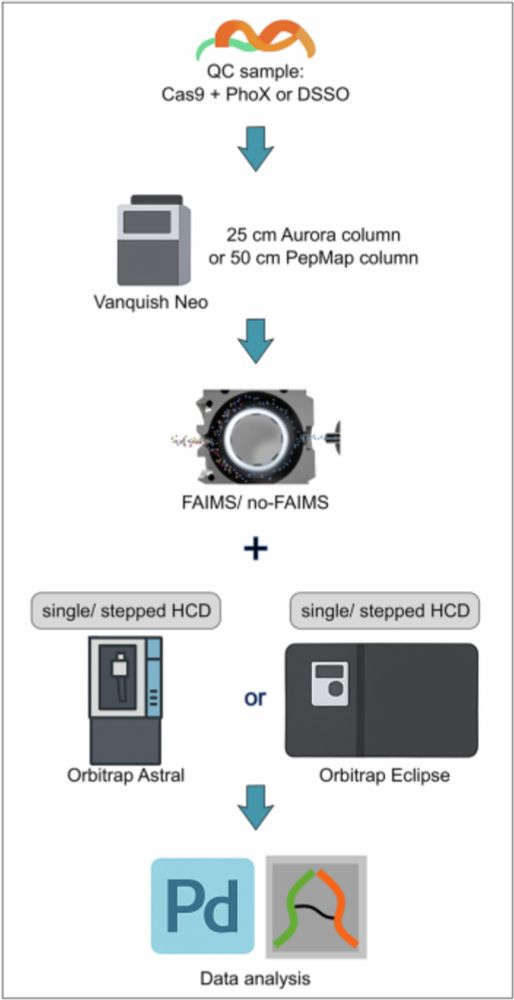
New paper alert! Researchers at the Proteomics Tech Hub at the @viennabiocenter.bsky.social have developed a new cross-linking mass spectrometry workflow that allows scientists to more easily detect and study protein interactions that were previously undetectable. More: bit.ly/4nV5n1w
12.11.2025 09:25 — 👍 8 🔁 2 💬 0 📌 0

GitHub - hadley/genzplyr: dplyr but make it bussin fr fr no cap
dplyr but make it bussin fr fr no cap. Contribute to hadley/genzplyr development by creating an account on GitHub.
Do you teach #rstats? Do your students complain about how lame and old-fashioned dplyr is? Don't worry: I have the solution for you: github.com/hadley/genzp....
genzplyr is dplyr, but bussin fr fr no cap.
06.11.2025 23:25 — 👍 460 🔁 168 💬 43 📌 56

Register now!
Five days of keynotes, workshops, flash talks, posters, and more on computational mass spectrometry
Confirmed keynote speakers
Armin Soleymaniniya · Arthur Declercq · Claire Koenig · Hannah Boekweg · Jonas Scheid · Karel Berka · Matthias Anagho-Mattanovich · Roman Bushuiev · Valdemaras Petrosius
📆 19-23 January 2026
📍 Harrachov, Czechia
Registration is now open for the EuBIC-MS Winter School 2026!
Join us in Harrachov, Czechia for a week of keynotes, workshops, and networking on computational MS.
Info and registration: eubic-ms.org/events/2026-...
#EuBIC2026 #MassSpectrometry #MassSpec #Bioinformatics #Proteomics #Metabolomics
16.10.2025 09:36 — 👍 15 🔁 11 💬 1 📌 2

Last week we had our yearly organizational meeting, this time in Vienna, Austria. We had 3 great days discussing the activities of the past year and planning the upcoming ones. Thanks to everyone who joined and contributed to a great meeting!
03.10.2025 11:33 — 👍 6 🔁 3 💬 0 📌 0
Also, I know that in DIA-NN there is a "threads" parameter that can be set, for the i9 that should be at least at 24 (and the OS hopefully schedules that to be each on separate cores). Maybe that helps :)
19.08.2025 22:37 — 👍 1 🔁 0 💬 1 📌 0
Haha pretty sure they'd be happy to hear about it though. As a dev I can confirm that it's hard to keep track of all the changes in the hardware world ;)
19.08.2025 22:36 — 👍 1 🔁 0 💬 0 📌 0
Might be because the software maybe isn't adapted to properly use all the cores in the new Intel CPUs yet? E.g. no more hyper threading on Intel CPUs and if it only uses performance cores there will be a huge performance loss compared to Ryzen CPUs. Probably worth asking the devs.
19.08.2025 20:20 — 👍 1 🔁 0 💬 1 📌 0
Excited to share our latest in vivo crosslinking workflow using a dual enrichment strategy: Over 5000 crosslinks and 56 novel PPls- Huge thanks to the amazing team: Philipp Bräuer, Laszlo Tirian, Fränze Müller, Karl Mechtler at the Vienna BioCenter who made this possible! doi.org/10.1038/s420...
16.08.2025 19:18 — 👍 13 🔁 2 💬 0 📌 0

In vivo crosslinking and effective 2D enrichment for proteome wide interactome studies www.nature.com/artic...
---
#proteomics #prot-paper
17.08.2025 09:20 — 👍 3 🔁 4 💬 0 📌 0
6th ESCP 2025 – APMA
The deadline to submit abstracts and register for this years symposium on single cell proteomics #ESCP25 in Vienna is approaching this Sunday! This year we additionally host a workshop for deep visual proteomics and a Thermo User Meeting.
25.07.2025 08:42 — 👍 7 🔁 3 💬 1 📌 0

Time to get signed up for the Symposium on Structural Proteomics! This time at @humantechnopole.bsky.social in Milan 6th-8th October Full speaker lineup announced. Thanks to @thermofishersci.bsky.social @brukercorporation.bsky.social, Affipro and MSVision for support
Info
ssp2025.squarespace.com
22.07.2025 15:12 — 👍 7 🔁 5 💬 0 📌 3

I would like to inform you that the registration deadline for our ESCP25 conference is July 27th.
We are looking forward to interesting scientific contributions.
10.07.2025 20:39 — 👍 12 🔁 7 💬 0 📌 0
‼️ Job alert 🚨: I have an open 3-year postdoc position in my lab to develop novel crosslinking mass spectrometry workflows. 🧪🧪 So if you want to do cool stuff in protein MS, please apply ! ⬇️ Please RT
www.verwaltung.uni-halle.de/dezern3/Auss...
#Academicsky
#Chemsky
#teammassspec
25.06.2025 12:44 — 👍 32 🔁 27 💬 0 📌 2

Redox proteomics in lung cancer patients reveals that there is a higher oxidation of the cysteines of glycolytic and methylglyoxal metabolism enzymes. doi.org/10.1038/s414...
17.06.2025 16:15 — 👍 4 🔁 1 💬 0 📌 0

Conference stage with Marie Locard-Paulet, the chairs, and the opening slide in the background.
Marie Locard-Paulet presenting the EuBIC-MS #ProteoBench project at the #EuPA2025 conference.
👉 Learn more about our open proteomics software benchmarking platform at proteobench.readthedocs.io.
#Proteomics #Bioinformatics #Benchmarking
19.06.2025 12:13 — 👍 19 🔁 5 💬 0 📌 1
🚧 Refactored lesSDRF; feedback welcome!
I’ve pushed a new version of lesSDRF to make high-throughput SDRF annotation faster and more intuitive.
Github issue: github.com/CompOmics/le...
Temporary app: refactorlessdrf.streamlit.app
I could use some feedback before making this the new lesSDRF 🙏
20.05.2025 14:25 — 👍 5 🔁 2 💬 1 📌 0
And big thanks also to the @eubic-ms.org hackathon team contributing to our #SpectriPy package github.com/RforMassSpec... !
Next on the list submission of the package to 👉 @bioconductor.bsky.social
11.04.2025 10:56 — 👍 14 🔁 5 💬 0 📌 0
Screenshot of the new Altmetric Explorer integration of Thoughts.
#AcademicSky #HigherEd #PsySky #Brain #Machine #Interfaces
THREAD
Today Altmetric is announcing a major development in collaboration with the Brain Occipital Lobe Learning Observation Centre Kalhausen-Strasbourg.
From today Altmetric will be able to determine what research paper you're thinking of, regardless of what you're actually writing/saying.
1/17
01.04.2025 06:10 — 👍 175 🔁 41 💬 7 📌 40
Privacy-first AI assistant by @proton.me, built in Europe. Lumo does not track or record your conversations.
Ask me anything, it's actually confidential.
"Cesar Nombela" Talento Fellow. Quantitative proteomics to unravel post-translational regulation. Currently at CNIC (Madrid) 💜🛠️👩🏻🔬🧫🔬
Proteomes (ISSN 2227-7382) is an #openaccess, peer reviewed journal on all aspects of #Proteomics science, published quarterly online by @mdpiopenaccess.bsky.social
I like big data and I cannot lie | Informatics/ML lead @ Belharra Tx, wet lab PhD @ Cravatt Lab
Ass. Prof., analytical chemist, mother, chess player, loves mass spectrometry and diversity in life as well as analysis
Pastel BioScience | t.ly/y4tAy
Proteomics conferences here >>> t.ly/GSBz-
Proteomics databases and scripts here >>> t.ly/iNYXg
Assistant Professor at the University of L'Aquila, Italy.
Scientist - Structural Proteomics, Mass spectrometry at Human Technopole. Views are my own. #XL-MS #TeamMassSpec #StructuralProteomics #StructuralBiology #Proteomics
Staff Scientist in Mass spectrometry platform @fmp-berlin.de. Interested in cross-linking mass spectrometry and interactomics.
Der Wissenschaftsfonds FWF ist Österreichs zentrale Einrichtung zur Förderung der Grundlagenforschung.
www.fwf.ac.at
scilog – das Wissenschaftsmagazin des FWF:
scilog.fwf.ac.at
Group Leader and Adjunct Professor 🧑🏫 | Public Health | Data Engineering & Visualization | 🧬 Bioinformatics & 📈 Mass Spec Enthusiast | 📍 Berlin | Passionate About Collaboration and Open Science 🔬✨
Offizieller Account der Uni Wien / Official account of University of Vienna.
Es postet http://communications.univie.ac.at 💬
Impressum: http://go.univie.ac.at/impressum Netiquette: http://go.univie.ac.at/netiquette
Research, Education and Business. Vienna BioCenter is a life science cluster that includes IMP, IMBA, GMI, Max Perutz Labs, as well as the Faculty of Life Science & CeMESS (both University of Vienna) and 35+ biotechs.
EMBL's European Bioinformatics Institute (EMBL-EBI) provides open biological data resources and tools, and performs basic research in computational biology. https://www.ebi.ac.uk/
Biologist and proteomics specialist at Max Perutz Labs Vienna.
Proteomics | mass spec | scientific illustration | Magic: the Gathering
Group leader at Leibniz-Forschungsinstitut für Molekulare Pharmakologie
W2-S Professor at Charité – Universitätsmedizin Berlin