Check out our new review on Graph #NeuralNetworks in #MolecularDynamics!
We show how #AI is used for #ForceField development, free-energy, and analysis - including our attention-based GNN workflow - and how you can integrate your favorite AI into #compchem!
www.sciencedirect.com/science/arti...
25.02.2026 23:46 —
👍 9
🔁 2
💬 0
📌 0

Want to know everything about #ArtificialIntelligence, #DeepLearning, and #CompChem to study and engineer #GeneEditing systems — from #CRISPR-Cas proteins to base and prime editors, and #RNA Guided Transposons?
Check out our new piece in @NatureSMB !
www.nature.com/articles/s41...
17.02.2026 19:01 —
👍 9
🔁 2
💬 0
📌 0
Thrilled to share that I’ll be joining @uclacb.bsky.social as Professor in July 2026!
Grateful to my amazing students, postdocs, colleagues @UCR, collaborators, and mentors.
A new chapter begins, advancing #compchem, #AI to explore #RNA, #DNA, #CRISPR, and emerging directions in #Biophysics.
31.10.2025 06:34 —
👍 20
🔁 3
💬 1
📌 0
Developed by the multi-talented @pabloarantes.bsky.social & @SouvikSinha members of my lab:
ParametrizANI brings #MachineLearning - based #ForceField parametrization available to everyone
Available on Github: github.com/palermolab/P...
09.10.2025 19:44 —
👍 9
🔁 1
💬 0
📌 0

🚀 Many thanks to @SDSC_UCSD and @HPCwire for highlighting our work using @NSF supercomputers to uncover how “jumping genes” can transform #GeneEditing technologies!
www.sdsc.edu/news/2025/PR...
#Supercomputing #HPC #CRISPR #RNA #CompChem
22.09.2025 19:45 —
👍 7
🔁 1
💬 0
📌 0

✨Best way to end the week! Just learned from Kevin Freedman @ucriverside.bsky.social that, at the end of the exam, students in his Dynamics of Biological Systems class draw their favorite professor.
Guess who got picked...
♥️♥️♥️
#biophysics #TeachingJoy #STEM #LoveScience
29.08.2025 22:50 —
👍 8
🔁 1
💬 2
📌 0
This is work mainly done by Mohd Ahsan, an incredibly talented postdoc in the lab. Grateful for the experimental validation by David Taylor lab @utaustin.bsky.social and NMR by George P. Lisi lab @brownuresearch.bsky.social
27.08.2025 17:48 —
👍 5
🔁 0
💬 0
📌 0

🚀New from our lab @biorxivpreprint.bsky.social!
We reveal a cryptic metal binding pocket that dinamically forms in #CRISPR-Cas9 and regulates the metal-dependent activity of #Cas9.
#MarkovModels & #QuantumChemistry corroborated by biochem & #NMR experiments.
www.biorxiv.org/content/10.1...
27.08.2025 17:45 —
👍 14
🔁 1
💬 1
📌 0
@pabloarantes.bsky.social is continuing working on improving the tool
21.08.2025 04:09 —
👍 1
🔁 0
💬 0
📌 0
Developed by the incredbly creative @pabloarantes.bsky.social & @SouvikSinha in our lab!
20.08.2025 17:33 —
👍 1
🔁 0
💬 0
📌 0

GitHub - palermolab/ParametrizANI: ParametrizANI - Fast, Accurate and Free Dihedral Parametrization in the Cloud with TorchANI
ParametrizANI - Fast, Accurate and Free Dihedral Parametrization in the Cloud with TorchANI - palermolab/ParametrizANI
New from our lab!
🚀 ParametrizANI - a #NeuralNetworks tool for molecular parametrization.
Predicts potential energy surfaces with near-DFT/CC accuracy, at a fraction of the computational cost!
#AI #QuantumChemistry #CompChem #ML 🤖
chemrxiv.org/engage/chemr...
Try it:
github.com/palermolab/P...
20.08.2025 17:33 —
👍 37
🔁 9
💬 3
📌 0


So exciting to be @proteinsociety.bsky.social meeting with my lab!
29.06.2025 18:53 —
👍 7
🔁 0
💬 0
📌 0
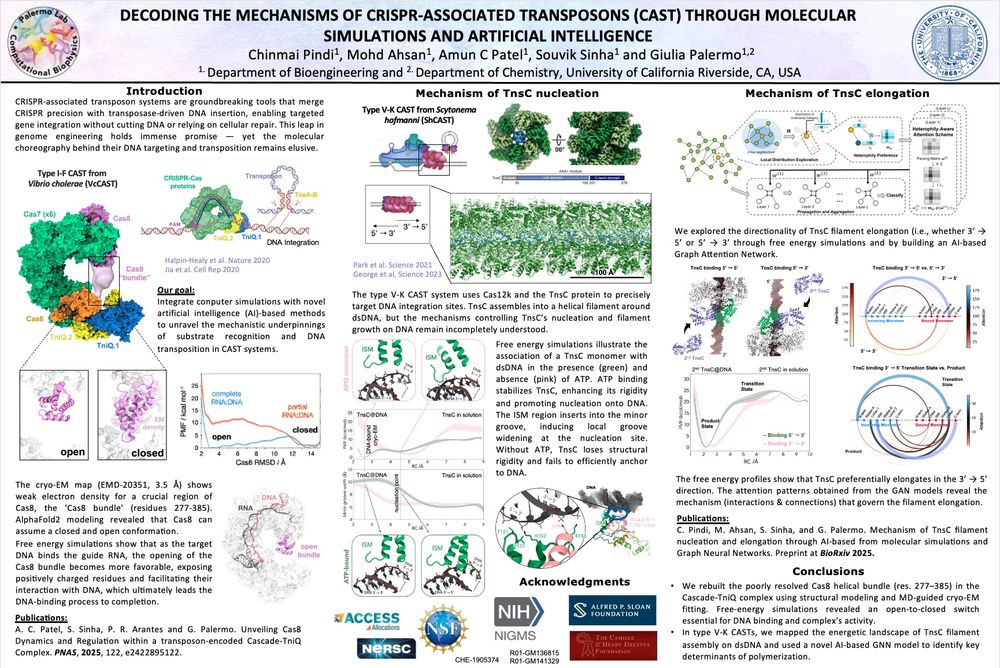
🔬🧬 Excited to be at the @proteinsociety.bsky.social meeting in SF! Curious how #NeuralNetworks can decode biomolecular mechanisms? Don’t miss the poster by ChinmaiPindi and AmunPatel ! #Proteins #AI #CRISPR #DNA
28.06.2025 20:42 —
👍 9
🔁 0
💬 0
📌 0
This was developed by Chinmai Pindi, Mohd Ahsan and Souvik Sinha, Ph. D, three amazing lab members! More on #NeuralNetworks, #GNN & new #AI approaches is yet to come! Stay tuned !
19.06.2025 20:13 —
👍 2
🔁 0
💬 0
📌 0
By leveraging deep learning–based graph representations, our GAT model provides interpretable mechanistic insights from complex molecular simulations and is readily adaptable to a wide range of biological systems.
19.06.2025 20:10 —
👍 2
🔁 0
💬 0
📌 0
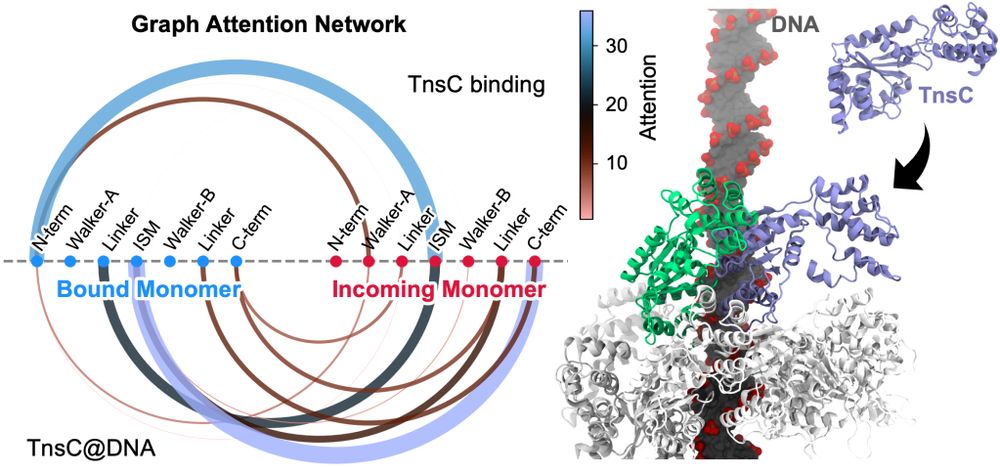
New study @biorxivpreprint
We revea, how TnsC filaments form and grow in #CRISPR-associated transposons using #FreeEnergy sims & introduce #DeepLearning-based Graph Attention Networks for decoding biomolecular systems!
www.biorxiv.org/content/10.1...
#GNN #MolecularDynamics #StructuralBiology
19.06.2025 20:09 —
👍 10
🔁 1
💬 2
📌 0
New paper from our lab just out @pnas.org! 🚀
Using free energy simulations, #AIphaFold and #cryoEM refinement, we uncovered a key conformational change in a paradigm-shifting #CRISPR-associated #transposon capable of precise gene knock-ins. 🔍🧬
www.pnas.org/doi/epub/10....
02.04.2025 16:28 —
👍 29
🔁 5
💬 0
📌 1
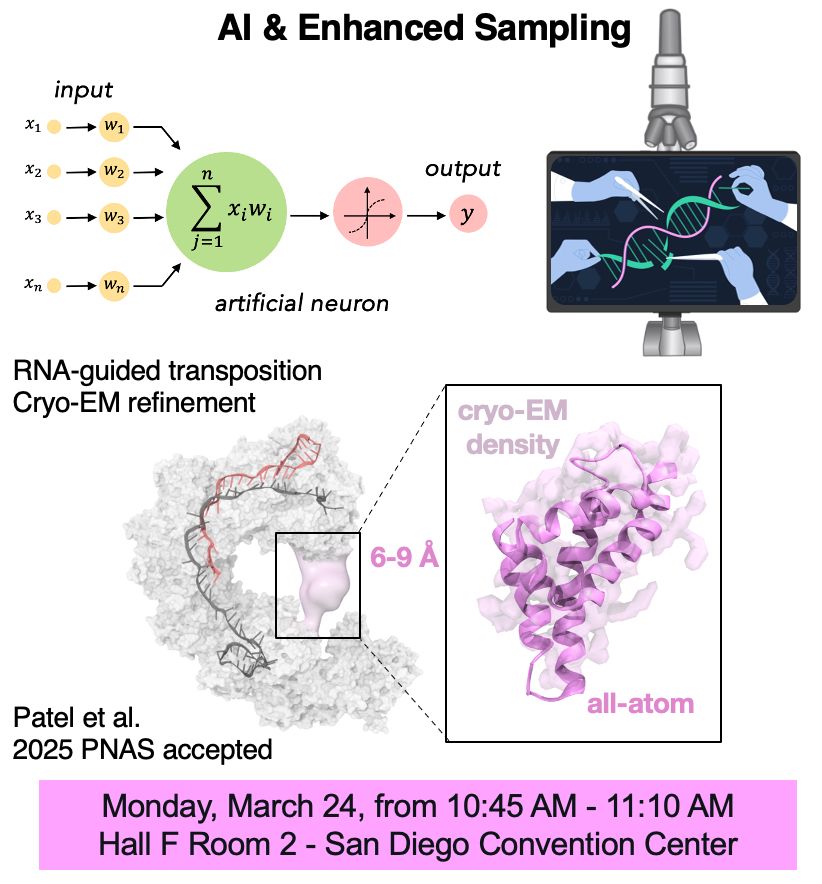
Thrilled to join #ACSSpring2025 and meet my friends! Don't miss my talk on how #AI & #EnhancedSampling can describe gene editing & refine #cryoEM maps. We’ll be presenting our latest research on #CRISPR-associated transposon systems + other exciting advancements! #compchem #RNA
#compchem #RNA
23.03.2025 19:33 —
👍 10
🔁 1
💬 0
📌 0

#BPS2025 waiting for the national lecture 🥳 @giuliapalermo.bsky.social @labvanni.bsky.social @sriraksha.bsky.social @sesamedusa.bsky.social
18.02.2025 04:01 —
👍 16
🔁 2
💬 2
📌 0
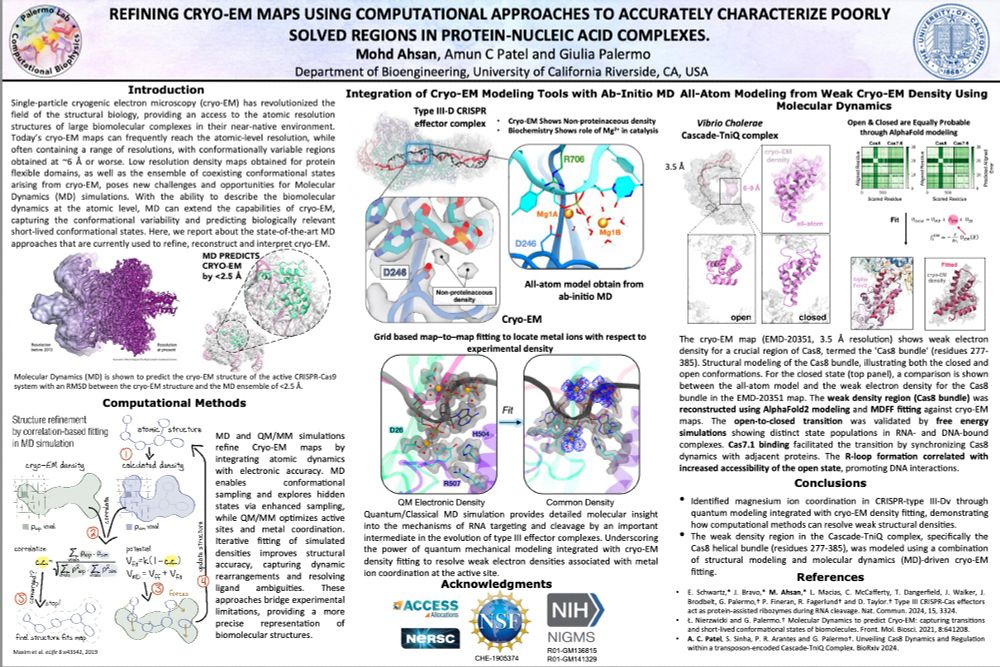
🚀 Exciting new poster from our lab at #BPS2025!
🧬 @Ahsan09966804 showcases how #MolecularDynamics & #AI can enhance #CryoEM map refinement.
📍 Catch it at Board 18 (1824-Pos) today, don’t miss out! #Biophysics #CompBio
17.02.2025 18:15 —
👍 16
🔁 2
💬 0
📌 0
Carolina Sarto @carosarto.bsky.social, a brilliant student from the University of Buenos Aires, will be presenting her work on #RNA synthesis in #DengueVirus🦠 at #BPS2025.
Don’t miss her poster today L3074-Pos (LB13). She’s looking for a postdoc, and I have no doubt she’ll thrive!
16.02.2025 20:19 —
👍 7
🔁 1
💬 1
📌 0
@carosarto.bsky.social
16.02.2025 20:15 —
👍 1
🔁 1
💬 0
📌 0
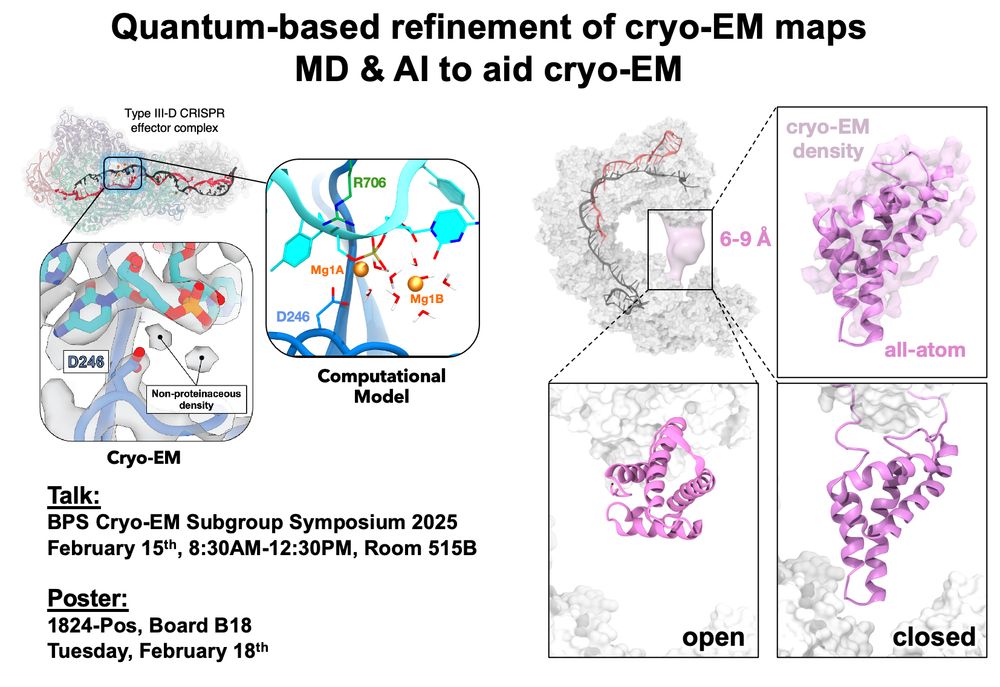
Excited to be at #BPS2025! 🚀 Our lab is contributing to the #cryoEM symposium, showcasing how computational methods and AI are revolutionizing cryo-EM analysis. See you there! 🔬💡 #Biophysics #AI
Read our papers to know more:
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...
15.02.2025 15:11 —
👍 25
🔁 4
💬 0
📌 0
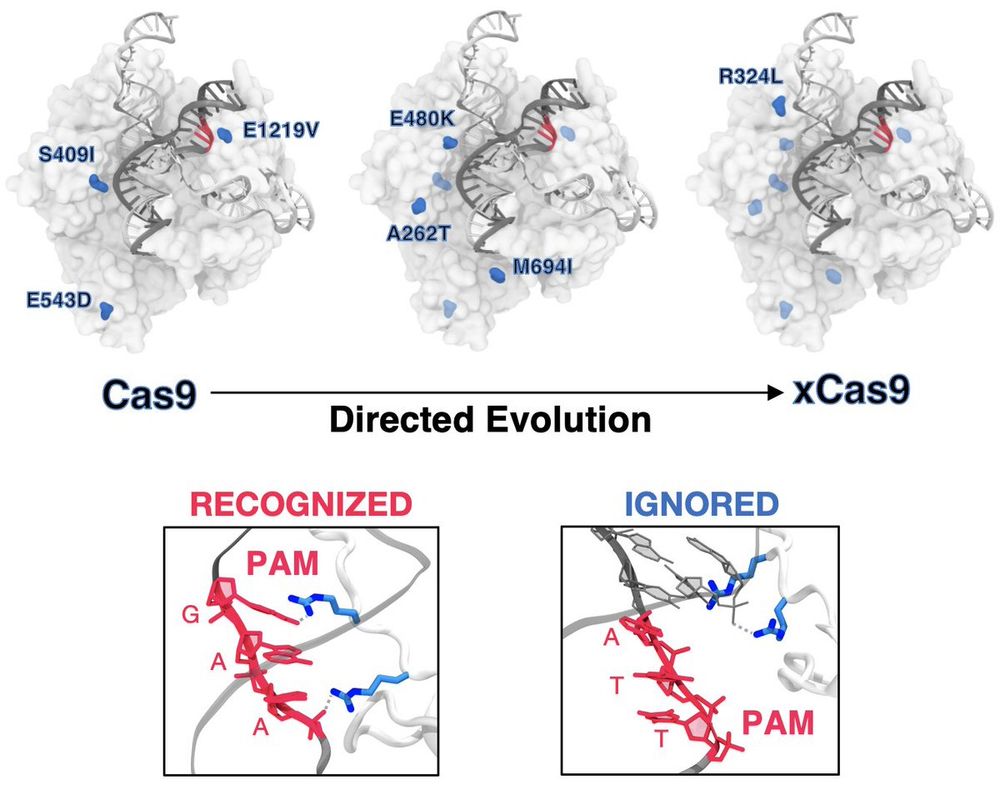
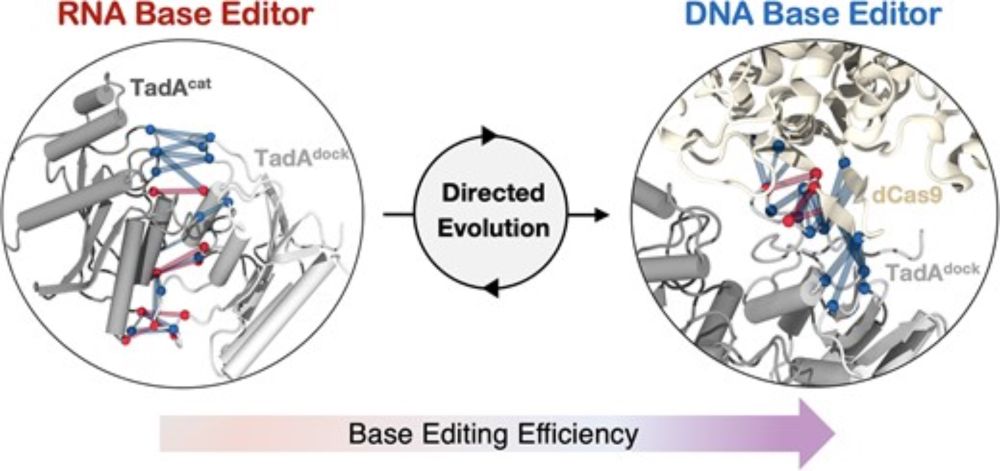
New paper from our lab @elife.bsky.social . We show how #DirectedEvolution reshaped #CRISPR for expanding DNA recongition.
This is work by Kazi Hossain, an incredibly talented student visiting my lab from the Czub lab, now in Orozco's lab & Lukasz Nierzwicki.
doi.org/10.7554/eLif...
11.02.2025 08:22 —
👍 9
🔁 2
💬 0
📌 0
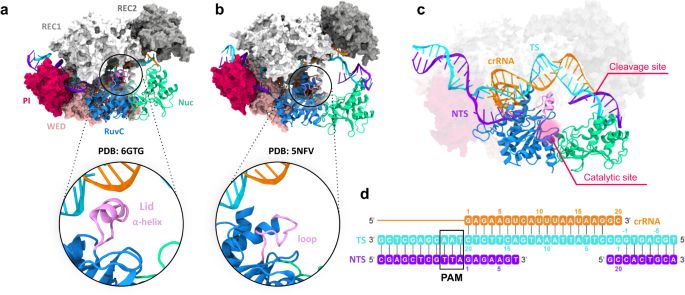
New paper from our lab @naturecomms.bsky.social!
We reveal the dynamics and mechanism of target DNA traversal in #CRISPR Cas12a, a conundrum in the field!
nature.com/articles/s41...
#compchem
We thank the amazing #HPC resources of PSC #Anton2 and SDSC
08.02.2025 18:22 —
👍 45
🔁 9
💬 3
📌 0
Happy Bluesky! Nel blu dipinto di blu 🩵🩵🩵
08.02.2025 04:48 —
👍 4
🔁 2
💬 0
📌 0