🚀We’re hiring! Join our international team at @ciid-heidelberg.bsky.social and @uniheidelberg.bsky.social
as a PhD Student or Postdoctoral Researcher to lead cutting edge research projects at the virus-host interface
Postdoc: tinyurl.com/d5sdhbnp
PhD: tinyurl.com/bdz5e5u6
01.10.2025 13:38 —
👍 5
🔁 4
💬 0
📌 0
Thanks a lot Marco!
11.07.2025 16:51 —
👍 1
🔁 0
💬 0
📌 0
Thanks a lot Stefan!
11.07.2025 16:50 —
👍 0
🔁 0
💬 0
📌 0
We’re gearing up to grow the Heidelberg team and will soon post several exciting positions. Stay tuned! 4/4
01.07.2025 13:31 —
👍 3
🔁 0
💬 0
📌 0
A huge thank you to my mentors, colleagues & friends, and especially my team - your hard work got us here. Also, many thanks to the broader virology community; it’s a privilege to contribute to such an amazing and dynamic field. 3/4
01.07.2025 13:31 —
👍 3
🔁 0
💬 1
📌 0
I’m excited to take on this new task and look forward to working alongside the incredible teams at the Center for Integrative Infectious Disease Research (@ciid-heidelberg.bsky.social) and across the Heidelberg Medical Campus. 2/4
01.07.2025 13:31 —
👍 2
🔁 0
💬 1
📌 0
Big personal news: Today I’ve started as Full Professor & Director of the Molecular Virology Department at Heidelberg University (@uniheidelberg.bsky.social). 1/4
01.07.2025 13:31 —
👍 21
🔁 2
💬 2
📌 2

© DORANTHPOST
The #HIRI & the Faculty of Medicine at the University of Würzburg jointly open a call for 3 HELMHOLTZ RESEARCH GROUPS WITH JUNIOR PROFESSORSHIPS (W1) in the areas of
🔹RNA-based Infection Research
🔹Microbial RNA Biology
🔹RNA-based Medicine
Apply by January 19 at www.helmholtz-hiri.de/en/w1-groups/
12.12.2024 12:41 —
👍 12
🔁 11
💬 1
📌 4

On the trail of RNA
A daring e-mail took biochemist Mathias Munschauer from Mannheim to New York, to the laboratory of one of the world’s most renowned RNA
A daring email took @munschauerlab.bsky.social from Mannheim to New York, to the lab of one of the world’s most renowned #RNA researchers. 👨🔬🧬 Today, he is one of the most sought-after experts in the field. Learn more about him in this portrait by @www.helmholtz.de: www.helmholtz.de/en/newsroom/...
28.06.2024 08:10 —
👍 8
🔁 2
💬 0
📌 0
In conclusion, SHIFTR enables the systematic mapping of region-resolved RNA interactomes for any RNA in any cell type and has the potential to revolutionize our understanding of transcriptomes and their regulation.
12.02.2024 08:09 —
👍 1
🔁 0
💬 0
📌 0
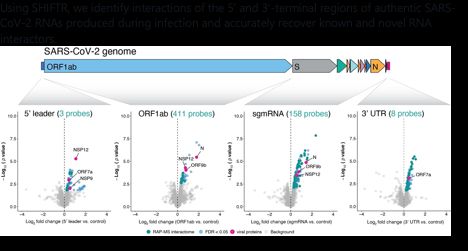
Using SHIFTR, we identify interactions of the 5′ and 3′-terminal regions of authentic SARS-CoV-2 RNAs produced during infection and accurately recover known and novel RNA interactors.
12.02.2024 08:09 —
👍 1
🔁 0
💬 1
📌 0
SHIFTR works for RNAs of different length and abundance (snRNAs, lncRNAs, mRNAs) and is compatible with sequentially releasing interactomes for multiple target RNAs in a single experiment.
12.02.2024 08:08 —
👍 0
🔁 0
💬 1
📌 0
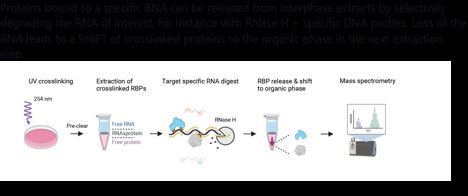
Proteins bound to a specific RNA can be released from interphase extracts by selectively degrading the RNA of interest, for instance with RNase H + specific DNA probes. Loss of the RNA leads to a SHIFT of crosslinked proteins to the organic phase in the next extraction step.
12.02.2024 08:08 —
👍 1
🔁 0
💬 1
📌 0
In brief, SHIFTR relies on the sequence-independent isolation of crosslinked RNA-protein complexes by organic phase separation as described in OOPS, XRNAX, or PTex.
12.02.2024 08:08 —
👍 1
🔁 0
💬 1
📌 0
With SHIFTR you can identify proteins directly bound to a specific region within any endogenous RNA. In addition to delivering region-resolution, SHIFTR requires orders of magnitude lower input material compared to the state-of-the-art, thus removing a critical bottleneck.
12.02.2024 08:07 —
👍 1
🔁 0
💬 1
📌 0
Time to SHIFT to BlueSky and share our recent paper introducing SHIFTR, a novel platform for region-specific RNA interactome discovery developed by PhD student Jens Aydin 👨🔬 in my group. Now published in Nucleic Acid Research: academic.oup.com/nar/advance-...
12.02.2024 08:07 —
👍 15
🔁 4
💬 2
📌 2
Big shout-out to the lab members leading this effort @Helmholtz_HIRI @Helmholtz_HZI: Nora Schmidt, Sabina Ganskih, Yuanjie Wei and Alex Gabel. We are immensely grateful to our collaborators and funders @ERC_Research and the Helmholtz Association.
04.10.2023 08:44 —
👍 1
🔁 0
💬 0
📌 0
In brief, the host protein SND1 binds the negative-sense RNA intermediates of SARS-CoV-2 and promotes viral RNA synthesis through recruitment of NSP9, which acts as a protein primer for RNA production 🤯.
04.10.2023 08:40 —
👍 2
🔁 0
💬 1
📌 0



