
Door with three posters, including Biology is Bigger than Binaries from SkypeAScientist, along with In this Lab we Believe (Science is real, love is love, black lives matter, feminism is for everyone, plants are cool, and immigrants are welcome)
Also a lab inside joke about poorly drawn dinosaurs from the movie "Drasik Werl" with a QR code that leads to the screeching recorder version of the Jurassic Park theme song.
New addition to the lab door courtesy of @sarahmackattack.bsky.social and squidfacts.bigcartel.com
29.05.2025 15:02 — 👍 135 🔁 19 💬 3 📌 1

Part II: T. tiliaceum in the elevator
01.04.2025 04:19 — 👍 0 🔁 0 💬 0 📌 0

We described this new species today: Orobanche andryalae, from the Canary Islands.
We described this new species today: Orobanche andryalae, from the Canary Islands. Welcome to science you beautiful thing ✨
20.02.2025 21:31 — 👍 199 🔁 38 💬 8 📌 0
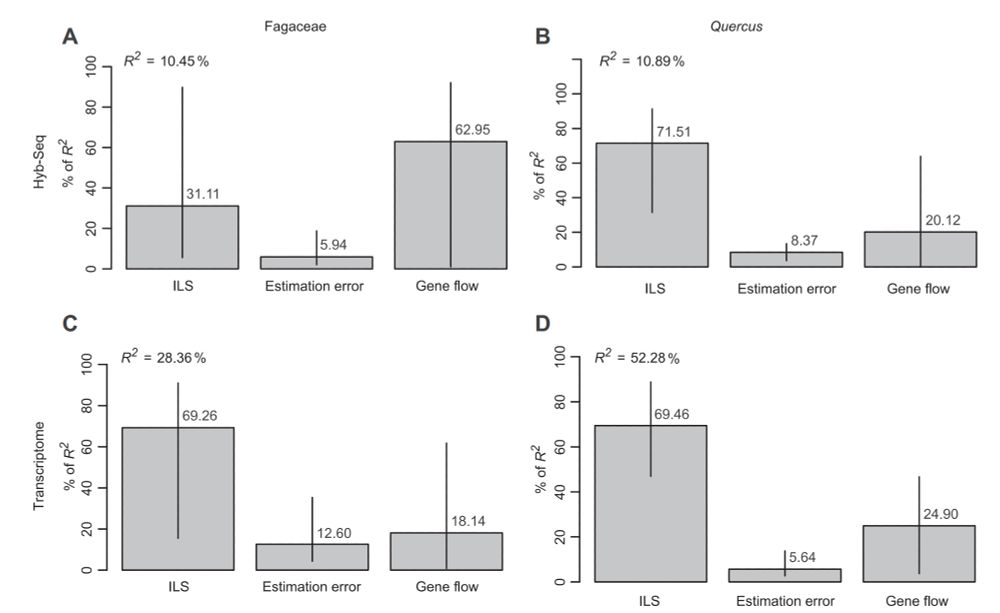
Figure 3. Plots showing the relative contributions of incomplete lineage sorting (ILS), gene tree estimation error, and gene flow to the
observed gene tree discordance, across two datasets and two taxonomic levels
The Hyb‐Seq (A–B) and transcriptome (C–D) datasets are represented here by the HYB‐98RT and RNA‐2821RT datasets, respectively. R2 denotes the total
proportion of gene tree variation explained by the model. Relative importance with 95% bootstrap confidence intervals is decomposed with the “lmg”
method and regressors of log transformations and summed to 100%.
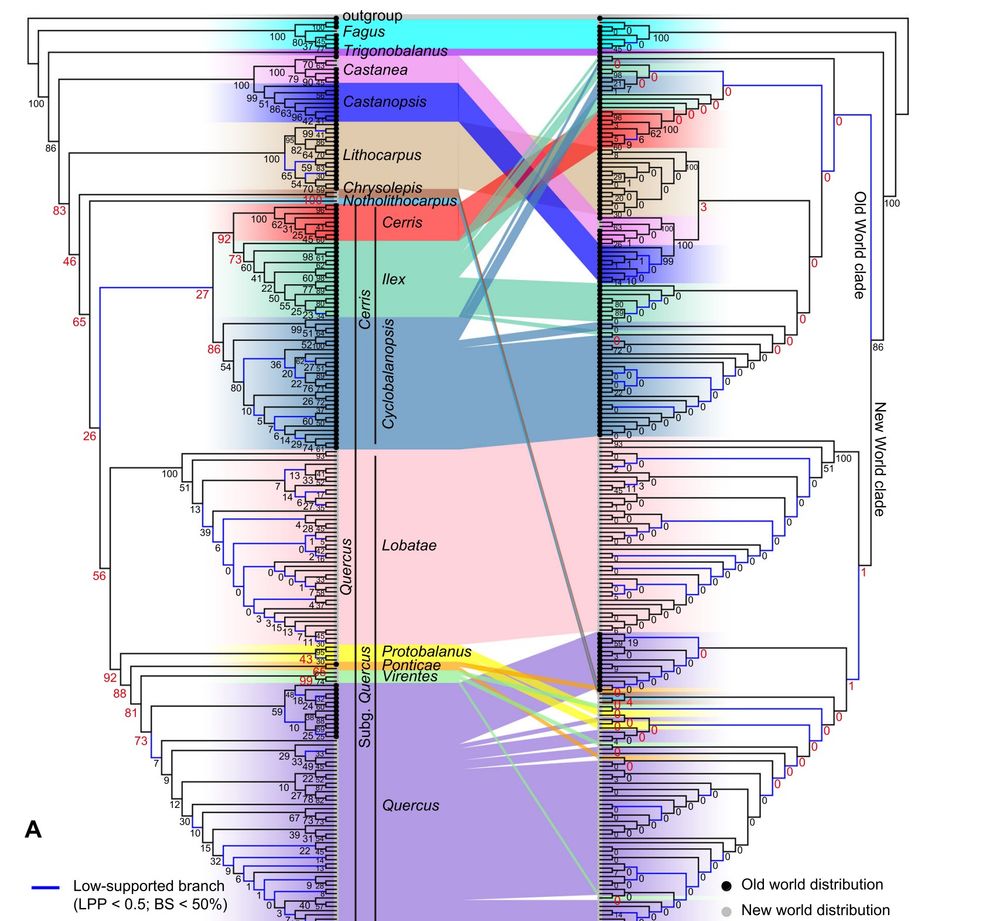
Figure 2. Discordance between nuclear and chloroplast phylogenies for oaks and relatives
(A) Cophylogeny showing incongruence between the nuclear ASTRAL (left; HYB‐98RT) and unpartitioned chloroplast maximum likelihood (ML) (right) trees.
Tip labels are shown in Figure S15. Clade frequencies of gene trees from the coalescent simulations are shown near the nodes; clade frequencies
associated with deep cytonuclear discordances are red and enlarged.
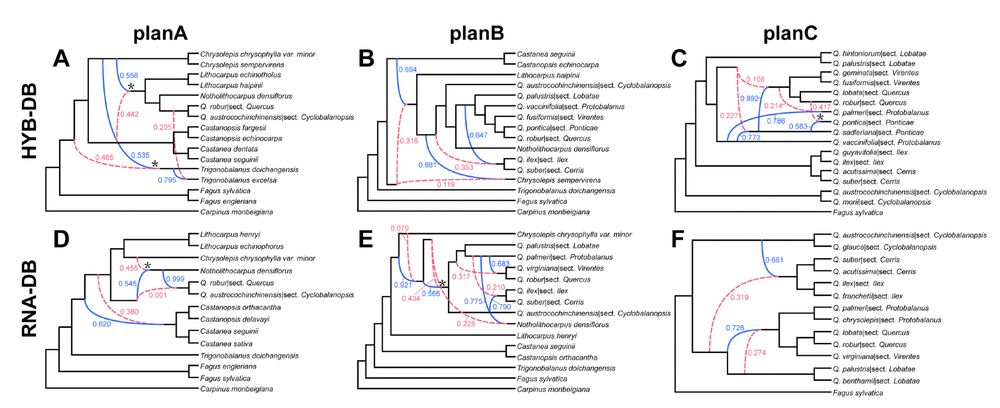
Figure S21. Species networks from reduced-representation datasets of oaks and Fagaceae. A. Optimal network under the “planA” taxon set of Fagaceae using the Hyb-Seq dataset (represented by the HYB-114RT dataset here). B. Optimal network under the “planB” taxon set of Fagaceae using the Hyb-Seq dataset. C. Optimal network under the “planC” taxon set of oaks using the Hyb-Seq dataset. D. Optimal network under the “planA” taxon set of Fagaceae using the transcriptome dataset (represented by the RNA-4853RT dataset here). E. Optimal network under the “planB” taxon set of Fagaceae using the transcriptome dataset. F. Optimal network under the “planC” taxon set of oaks using the transcriptome dataset. Dashed red curved branches denote the minor edge of a reticulate node while solid blue curved branches denote the major edge of a reticulate node. Values next to curved branches denote the inheritance probabilities. A reticulate node with an asterisk denotes that its parents contributed relatively equivalent inheritance probabilities (~ 0.5 vs. 0.5). Abbreviations: sect. CY, sect. Cyclobalanopsis.
Fascinating phylogenomic study of Oak Family by Shui‐Yin Liu et al. (including @carolsiniscalchi.bsky.social) showing that hybridization & ILS both generate gene-tree discordance in the family; ILS contributes ~3x as much as hyb'n to discordance in Quercus, but both matter.
doi.org/10.1111/jipb...
27.12.2024 04:23 — 👍 34 🔁 9 💬 1 📌 0

Casi 1K personas participaron en la elección de la especie amenazada 2025: Gracias 🙏
Resultó ‘Geranium dolomiticum’ que se enfrenta a presiones que tb limitan la interacción de polinizadores
Únete a la campaña de protección compartiendo este mensaje
♥️ #SEBICOP
#Conservación
#PlantaAmenazada2025 💚
23.12.2024 20:43 — 👍 30 🔁 12 💬 0 📌 0
Yeah not super great that a journal dedicated to ecology is happily using AI art when the environmental costs of generative AI is now a well known, widespread issue 🫠
01.12.2024 12:11 — 👍 453 🔁 115 💬 29 📌 4

Last Sunday I went to the shopping mall to take a round and I didn't expect to see my target species 🤣
26.11.2024 11:35 — 👍 1 🔁 0 💬 1 📌 0
Botany, biogeography, evolution, islands, photography.
Postdoc at National Museum of Natural History (Smithsonian Institution)
https://sites.google.com/view/albertojcoello
The goal of the Canadian BioGenome Project is to produce high-quality reference genomes 🧬 for all Canadian species 🌎
Sequencing Canada's Biodiversity 🌿🦋🐍🧬🐢🦌🐸🌳🐿🐙🦈🦦
Learn more: https://linktr.ee/canadianbiogenome
The Earth BioGenome Project (EBP), a moonshot for biology, aims to sequence, catalog, and characterize the genomes of all of Earth's eukaryotic biodiversity over a period of ten years.
🌲Keep up with all EBP updates: https://linktr.ee/earthbiogenomeproject
Associate Professor and researcher (Botany area) at Universidad Pablo de Olavide (Seville, Spain) and lifelong naturalist. #Iambotanist
Un árbol 🌲una vida. Por los bosques autóctonos y por la creación de un nuevo parque nacional.
Nature Plants is an online-only, monthly journal publishing the best research on plants (hopefully).
Cacereño. Físico y Oceanógrafo. Estudiante de doctorado en la UIB, investigando como titulado superior en la UMA.
The home of Kew Science 👩🔬
🌱Collections
📚Archives
🍄Applied research
🤝Partnerships
🥼Training
👉More: linktr.ee/kewscience
Spammers get blocked. | Gothic gardener. 🇨🇦. Goth, goblincore, dark art, horror, Halloween, poisonous plants. Nyctophile. Autistic murder hornet. Genderqueer. Pan. They/them. 🌿🍁🍂🎃
Interested on how plants evolve everyday, every season, every million years...
Ecology, pollination, networks, rstats & other personal stuff...
www.bartomeuslab.com
Ecologist working with species interactions, community dynamics and biodiversity maintenance 🌺🐝🌿
📍Berlin, Germany
Tropical ecologist | restoration | vegetation science | biodiversity monitoring | forests & grasslands | Amazonia & Andean mountains #BMAP #Smithsonian #DRYFLOR. ENG/SPA posts are mine
https://nationalzoo.si.edu/about/staff/reynaldo-linares-palomino
We are the British Ecological Society Macroecology & Macroevolution Special Interest Group. We run the annual #BESMacro conference and will gladly repost papers and opportunities of interest to our members!
Revista decana de la prensa ambiental. Observación, estudio y defensa de la naturaleza https://www.revistaquercus.es/
The Missouri Botanical Garden is an oasis in the city of St. Louis and a world leader in plant science, conservation, education and horticultural display.
Gardener horticulturist former Director of Hort for Royal Botanic Garden Sydney, CEO of Red Butte Garden and CityParklands of Brisbane. Plant geek & cook. Now just vegetable gardening in Northern New Zealand. 🏳️🌈
Associate Professor of Biology at Oxford University; plant chaser; artist; author. Find me on Insta @illustratingbotanist
#PlantBiologist Full professor @BiologiaUS studying #polyploidy #carnations, #phenotypicplasticity #molecularecology #functionalgenomics
http://personal.us.es/fbalao/
















