IsoNet2 determines cellular structures at submolecular resolution without averaging https://www.biorxiv.org/content/10.64898/2025.12.09.693325v1
11.12.2025 11:32 — 👍 30 🔁 9 💬 0 📌 4
Happy to share our fun team-up with @centriolelab.bsky.social & @stearnslab.bsky.social to reveal the architecture of the mammalian centriole’s Distal Ring 🛟. Very cool multiscale integration of #UExM 📏🔬with #CryoET ❄️🔬. Congrats to @ebertiaux.bsky.social & @computingcaitie.bsky.social #TeamTomo 🧪
10.12.2025 11:02 — 👍 51 🔁 18 💬 0 📌 0
CryoET of microbes inside an animal organ? Yes it’s possible! Check out our new preprint showing how we did it! What an amazing collaboration with amazing scientists who made this possible!
01.12.2025 04:35 — 👍 123 🔁 35 💬 2 📌 4
Prohibitins aren’t hanging out just anywhere in the membrane — they prefer the 💁♀️💎✨VIP section💅🪩🥂
Lab all-star @mmedina300kv.bsky.social maps these microdomains using cryo-ET + surface morphometrics.
s/o to #teamtomo #cryoET co-authors
@hamid13r.bsky.social
@attychang.bsky.social
@baradlab.com
17.11.2025 00:22 — 👍 54 🔁 17 💬 0 📌 2

Icecream: High-Fidelity Equivariant Cryo-Electron Tomography
Cryo-electron tomography (cryo-ET) visualizes 3D cellular architecture in near-native states. Recent deep-learning methods (CryoCARE, IsoNet, DeepDeWedge, CryoLithe) improve denoising and artifact cor...
Our colleagues Vinith Kishore and Valentin Debarnot from the @ivandokmanic.bsky.social lab have come up with an amazing deep learning tool for denoising and filling the missing wedge in #cryoET data. I'm pleased to introduce Icecream🍧
23.10.2025 11:12 — 👍 51 🔁 20 💬 3 📌 4
Hey #TeamTomo …would you like to try some 🍦ICECREAM!? Our @unibas.ch colleagues over at the @ivandokmanic.bsky.social lab have developed a stunningly effective tool to de-noise and de-wedge your tomograms. The contrast and details will give you brain freeze! 🍨❄️🤯
Please try, we need feedback 🧪🧶🧬🔬
23.10.2025 17:57 — 👍 44 🔁 16 💬 1 📌 1

Check out our new preprint on an integrated pipeline combining in situ #cryo-ET with MALDI #MSImaging for single-cell identification and classification from previously analysed EM-grids.
Link: www.biorxiv.org/content/10.1...
19.09.2025 18:07 — 👍 55 🔁 14 💬 1 📌 2

Amyloid reconstructions from cryoSPARC
Helical Reconstruction of Amyloids in cryoSPARC?
We present guidelines, limitations & future perspectives for processing amyloids in cryoSPARC @structurabio.bsky.social.
Great collaboration of the Kelly Lab and @landerlab.bsky.social #cryoEM #amyloid www.biorxiv.org/content/10.1...
05.10.2025 04:16 — 👍 44 🔁 14 💬 3 📌 0
Streptophyte algae go #TeamTomo with @thanahartmann.bsky.social!
02.10.2025 13:39 — 👍 5 🔁 0 💬 0 📌 0
Time for a thread!🧵 How different is the molecular organization of thylakoids in “higher” plants🌱? To find out, we teamed up with @profmattjohnson.bsky.social to dive into spinach chloroplasts with #CryoET ❄️🔬. Curious? ..Read on!
#TeamTomo #PlantScience 🧪 🧶🧬 🌾
elifesciences.org/articles/105...
1/🧵
25.09.2025 18:00 — 👍 135 🔁 42 💬 3 📌 6
Structural Analysis of Membrane Proteins in Cell-Derived Microvesicles
We have a preprint of a book chapter for you: Structural Analysis of Membrane Proteins in Cell-Derived Microvesicles.
We describe how to image membrane proteins in mid-sized vesicles using cryo-electron tomography
zenodo.org/records/1717...
22.09.2025 12:36 — 👍 51 🔁 17 💬 1 📌 0
These results are truly impressive! But I do worry that #cryoEM is moving into a phase where progress is made by optimizing parameter settings for black box programs. The devil is often in the details, and I am not sure how this could be implemented in #relion, as stated, without more info.
21.09.2025 07:42 — 👍 36 🔁 10 💬 2 📌 1

Sub-cellular chemical mapping in bacteria using correlated cryogenic electron and mass spectrometry imaging
Congrats Hannah Ochner and authors on this important paper! Strong collaboration with @kiranrpatil.bsky.social
www.biorxiv.org/cgi/content/...
@mrclmb.bsky.social @wellcometrust.bsky.social
31.08.2025 18:44 — 👍 142 🔁 62 💬 3 📌 4

If you are thinking about the future of structural biology, we are too! Our two cents (Jürgen Plitzko and I) just got published online here: authors.elsevier.com/sd/article/S...
23.08.2025 23:21 — 👍 89 🔁 27 💬 2 📌 0

Optimal tilt-increment for cryo-ET
Cryo-electron tomography (cryo-ET) enables high-resolution, three-dimensional imaging of cellular structures in their native, frozen state. However, image quality is limited by a trade-off between ang...
More great cryo-ET benchmarking work from my colleagues Maarten Tuijtel, Tomáš Majtner, Beata Turoňová, & @becklab.bsky.social: Systematic study of how tilt increment choice affects downstream TM & STA. Preprint has the optimally balanced tilt increment. #TeamTomo www.biorxiv.org/content/10.1...
23.08.2025 09:18 — 👍 32 🔁 7 💬 0 📌 2
🚨 Imagine a bacterium that refuses to follow the textbook:
It grows as tangled filaments, divides unevenly, reshapes its own membranes… and even builds grappling hooks.
Meet Litorilinea aerophila — and here’s why it blew our minds.
20.08.2025 10:34 — 👍 169 🔁 70 💬 5 📌 5
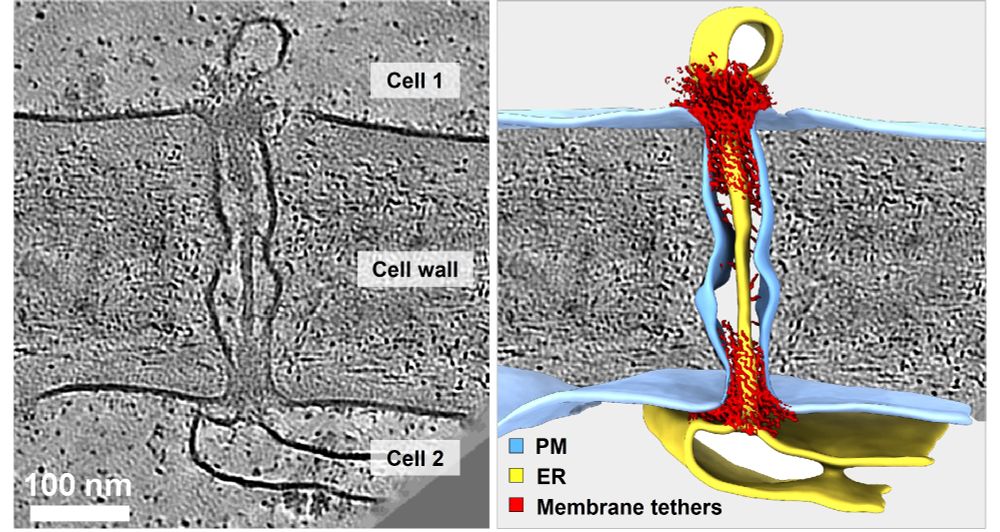
New preprint: In situ Architecture of #Plasmodesmata.
Using #cryoET, #AlphaFold & proteomics, we uncover the native organization of plant intercellular nanopores.
🔗 doi.org/10.1101/2025...
🧪🧵1/n
#teamtomo #PlantScience #Physcomitrium #Arabidopsis #cryoEM
26.07.2025 15:38 — 👍 196 🔁 71 💬 9 📌 8
hot (well, cold) off the press! ❄️
well done @glynnca.bsky.social & @michaelgrange.bsky.social & fabulous co-authors 👏🏼
#teamtomo #cryoET
21.07.2025 20:55 — 👍 10 🔁 1 💬 0 📌 0
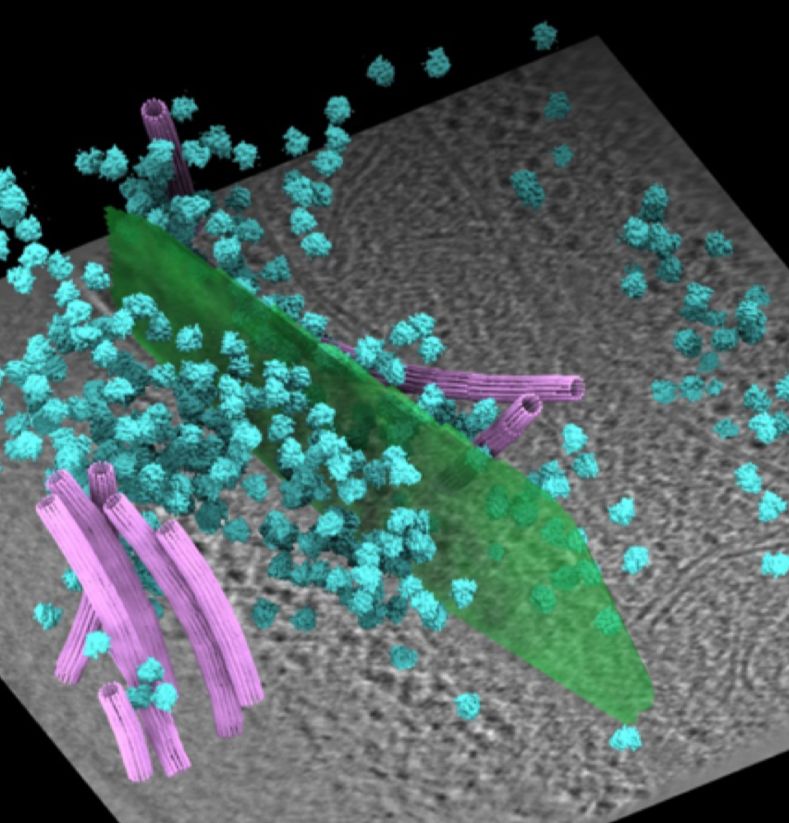
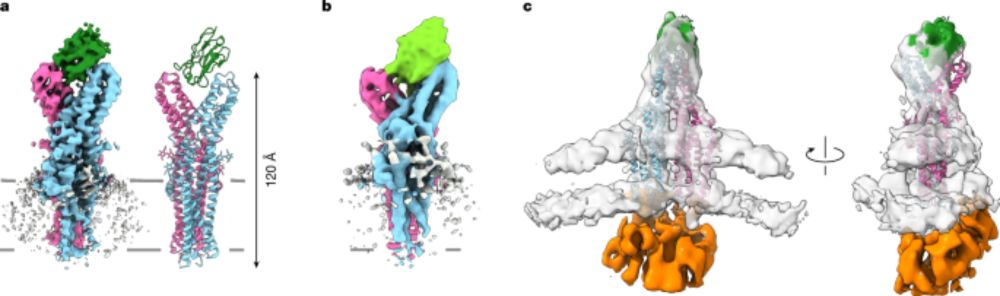
CryoET tomogram with the gap junction (green), ribosomes (cyan) and microtubules (pink)
In our latest preprint on bioRxiv, we present the in situ structure of
the human gap junction at 14 Å resolution using cryo-ET.
The structure provides a blueprint for investigating how connexin
regulation shapes intercellular communication in health and disease.
08.07.2025 05:33 — 👍 103 🔁 25 💬 4 📌 2
Delighted to see our work in press! 🙌🔬🧠
Thanks to @glynnca.bsky.social, @michaelgrange.bsky.social and the rest of the fabulous co-authors for all your contributions.
Excited to see where in tissue structural cell biology goes next 👀!
#liftlaughlove #teamtomo #whopper
19.06.2025 09:23 — 👍 8 🔁 3 💬 0 📌 0
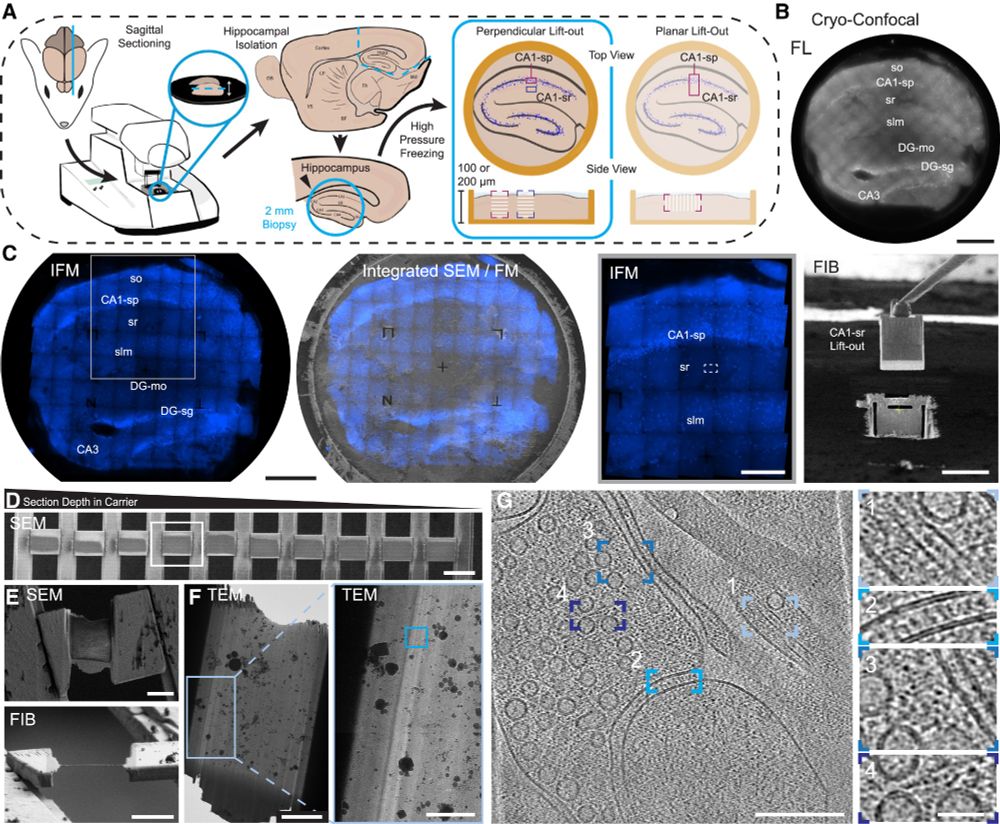
Proud to share our latest paper. doi.org/10.1016/j.cr...
Through the dedication of @glynnca.bsky.social and @cryingem.bsky.social we report a thorough method to image molecular organisation within hippocampus tissue.
Structural biology in tissue is well and truly here!
@rosfrankinst.bsky.social
17.06.2025 10:59 — 👍 62 🔁 24 💬 2 📌 4
Final PhD paper now reviewed and published in JSB:X. I was happy with some great reviews that improved the paper! Thanks to Sander Roet for his help with the code base, and Remco Veltkamp and @fridof.bsky.social
09.06.2025 06:56 — 👍 20 🔁 4 💬 0 📌 1
Proud to present the lab's latest work. The full structure of the sodium translocating methyltransferase (Mtr) bound to a small oxygen-responsive small protein MtrI. Work by Tristan and together with @schmitzstreitslab.bsky.social .
www.biorxiv.org/content/10.1101/2025.06.02.657420v1
04.06.2025 19:10 — 👍 58 🔁 22 💬 3 📌 3
We are happy to release LocScale2.0: a tool for context-aware, confidence-weighted cryoEM map optimisation.
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
22.05.2025 10:10 — 👍 121 🔁 38 💬 1 📌 2
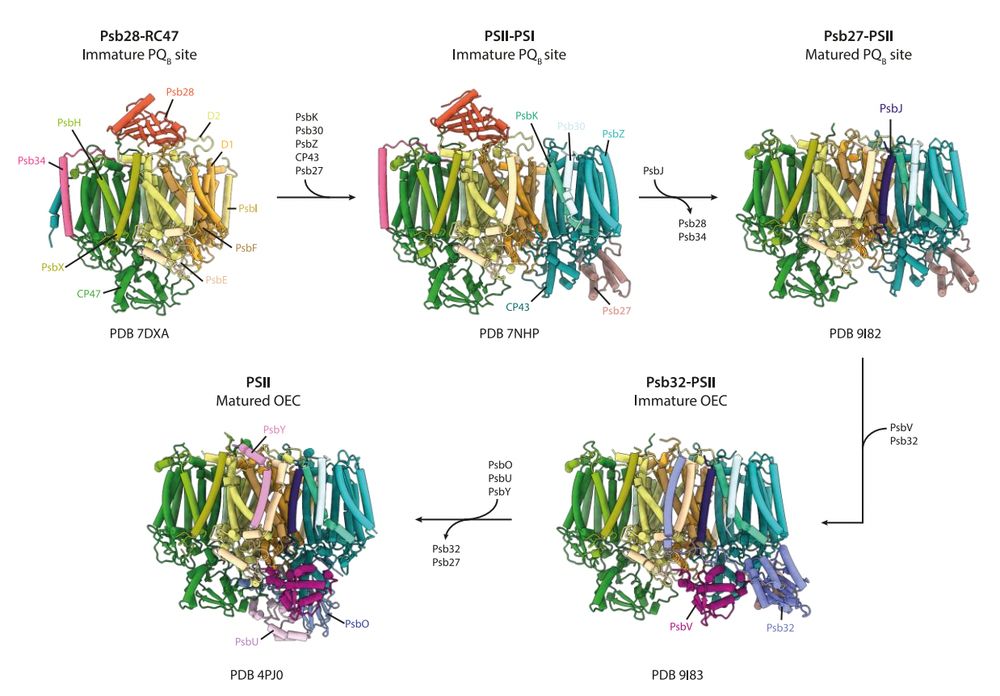
Happy to present the lab latest work, today from the green-side. The ultimate PSII assembly intermediate that directly precedes incorporation of the Mn4O5Ca cluster and formation of the OEC. Psb32 binds to PsbV, challenging the assumption the extrinsic subunits all associate spontaneously in-vivo.
12.05.2025 05:50 — 👍 37 🔁 14 💬 3 📌 2
Welcome to TomoGuide
A step-by-step Cryo-ET guide
Hey #TeamTomo,
Ever been in need of a tutorial about the fundamentals of cryo-electron tomography? From preprocessing raw frames to high-res subtomogram averaging?
That's why @florentwaltz.bsky.social and I made this website!
tomoguide.github.io
Follow the thread 1 /🧵
#CryoET #CryoEM 🔬🧪
06.05.2025 16:26 — 👍 116 🔁 41 💬 5 📌 7
Structural Biologist • Biochemist • Parasitologist • Co-host of ThePlungePodcast theplunge.org • Assistant Professor @ Columbia • Cryo electron microscopy/tomography of malarial membrane protein complexes. #teamtomo #cryoET #cryoEM #malaria cmholab.org
Post-Doc in Grotjahn lab (http://grotjahnlab.org) @scrippsresearch. Previously, PhD in Badrinarayanan lab (http://microgenomes.com) @NCBS_bangalore . 🏳️🌈
#mitochondria #cryoET
Director, Dept. of Mechanistic Cell Biology, Max Planck Institute of Molecular Physiology, Dortmund (DE). Opinions my own
The Young Scientific Symposium is a student-led scientific conference focused on the interface of chemistry and biology. Organized by PhD students and postdocs, the symposium brings together students and early-career researchers.
IECB - BORDEAUX
🇻🇳 structural biologist☃️cryo EM 🍖
group leader at IPBS Toulouse, France, ATIP Avenir group
Postdoc fellow at EMBL (DE)
"views are my own"☘️🌊🦠🧪🌈
Computational microscopist
#RNAcartography | #StructuralSystemsBiology | #Cryo-ET
More @ cryosagar.openemage.org
PhD student at Karolinska Institutet and Scilifelab | Cell physics
https://www.csi-nano.org/
PhD student at the University of Wisconsin-Madison Ané Lab.Interested in signalling pathways, symbiotic plant microbe interactions.
Assistant Professor @Aarhus University; interested in signalling pathways, symbiotic plant microbe interactions, nitrogen fixation
MAdLand is a DFG-funded research consortium exploring the molecular mechanism behind the transition from water to land, from alga to land plant.
Learn more on our Website: madland.science
PhD student at SYNMIKRO | Marburg 🇩🇪
Schuller Lab | #cryoET 🌱🔬
[aka Thana]
PhD student in Kudryashev lab MDC Berlin, cryoEM/cryoET, neurons, ion channels
Senior Cryo-EM Scientist, Astbury Biostructure Laboratory, University of Leeds
Postdoc at Imperial College
Bacterial conjugation & Cryo-EM
Staff scientist at the university of Osnabrück, exploring membrane transporters by #cryoEM
Assistant Professor of Biology at Boston University ::: Science Journalist / Communicator ::: National Geographic Explorer
MSCA post-doctoral researcher at Förster lab, Utrecht university.
SPA guy trying to navigate Tomo.
Lover of big protein complexes and even bigger scopes.
I'm new here. #structuralbiology #cryoem
M.D. | Physicist | Dr. rer. nat. | Investigating Tissue and Neurodegeneration by #CryoET, PostDoc @uniGoettingen, previous MPI Biochemistry
Environmental microbiologist, biogeochemical cycles, AMR in the environment
Royal Society Dorothy Hodgkin Research Fellow at UEA
Editor-In-Chief LAM Journal
🏳️🌈ally | mum of 2 | I love 🌋
www.marcelahernandez.com