
It’s official: ChatGPT can't draw a GPCR, but you’ll master them in the Filizola Lab
😜 Join Us! (send your CV and the names of at least two references to my institutional email) #Postdoc #scientist #research
@carlocamilloni.bsky.social
Associate professor at the University of Milano. Biophysics and computational structural biology. http://compsb.unimi.it Developer @plumed.org https://orcid.org/0000-0002-9923-8590

It’s official: ChatGPT can't draw a GPCR, but you’ll master them in the Filizola Lab
😜 Join Us! (send your CV and the names of at least two references to my institutional email) #Postdoc #scientist #research

Snapshot of a condensate near a lipid membrane with Swedish Research Council and Malmö University logos.
I'm hiring for a PhD position at Malmö University, Sweden!
The project will focus on molecular modelling of proteins, lipids, and biomolecular condensates at cell membranes.
More details and application form: tinyurl.com/4zm92365
Please feel free to share!
@vetenskapsradet.bsky.social | @mau.se
We asked what a strong static EF may do to AB42 fibril, we saw a marked perturbation of the fuzzy coat and increase fluctuations of the fibril core
27.09.2025 06:25 — 👍 2 🔁 0 💬 0 📌 0



Apple is entering the protein folding arena.
SimpleFold: Folding Proteins is Simpler than You Think arxiv.org/abs/2509.18480 🧬🖥️🧪 github.com/apple/ml-sim...
RFdiffusion3 is here! We train a general network that explicitly models every atom and use it to design active enzymes and DNA binders.
Tremendous team effort with Jasper, Rohith, Raktim, Rafi, Yanjing, Paul, Jonathan, and many others!
Check it out: lnkd.in/eiUFfJaM.

AtomWorks is out! Building upon @biotite_python, we built a toolkit for all things biomolecules and trained RF3 with it. All open-source, test it via `pip install atomworks`!
AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6

RosettaFold 3 is here! 🧬🚀
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...
🚀 Super excited to share our new work, led by @vschnapka.bsky.social bAIes, a Bayesian framework for generating accurate atomic-resolution ensembles of IDPs using #AlphaFold info! Now on @biorxivpreprint.bsky.social → feedback welcome! @pasteur.fr @erc.europa.eu
www.biorxiv.org/content/10.1...
the JIF 👇 is a very (!) narrow definition of "impact" = cites in first 2y after publication...
👉 What about "constant" or "late" impact?
👉 Why do we accept that a private company defines our impact?
👉 Why do we allow to be judged on this basis?
#researchintegrity #chemsky, #compchemsky

Rapid, nanoscale dynamics of individual IDPs determine bulk properties of the entire condensate, such as viscosity.
Amazing work by Nicola Galvanetto & the Schuler lab!
www.pnas.org/doi/10.1073/...
Congratulations to Michele Parrinello for receiving the ESI Medal 2025!
@esivienna.bsky.social
@univie.ac.at

A graphic for the Marie Skłodowska-Curie Actions (MSCA), showing a historical portrait of Marie Skłodowska-Curie overlaid with an image of four young researchers walking down a hallway. The European Commission logo is in the top left. Text reads: "Marie Skłodowska-Curie Actions – €404.3 million to support postdoctoral researchers”
Choose Science. Choose Europe.
A new Marie Skłodowska-Curie Actions Postdoctoral Fellowships 2025 call is now open.
With a budget of €404.3 million, it will support around 1,650 researchers from Europe and beyond.
Apply by 10 September → europa.eu/!fBTMgF
What an incredibly cool paper! While knot theory strictly applies to closed curves, Tommy, @smnlssn.bsky.social , and @paulrobustelli.bsky.social show that writhe, a knot "non-invariant" that changes with smooth deformations, provides a meaningful descriptor for flexible conformations.
01.05.2025 05:49 — 👍 7 🔁 2 💬 1 📌 0
onlinelibrary.wiley.com/doi/10.1002/... Federico
did some modeling to make some hypothesis on the interaction of anthocyanin with glucose transporter
Protein folding, binding and redox conditions by Stefano Gianni team in Rome the super productive Valeria Pennacchietti,
doi.org/10.1016/j.ij...
Very proud of Amit Kumawat, Elisa Tavazzani and all coworkers for this work on hERG K+ channel rescue mechanism by ICA. Now published here: jbiomedsci.biomedcentral.com/articles/10....
07.04.2025 10:27 — 👍 4 🔁 0 💬 1 📌 0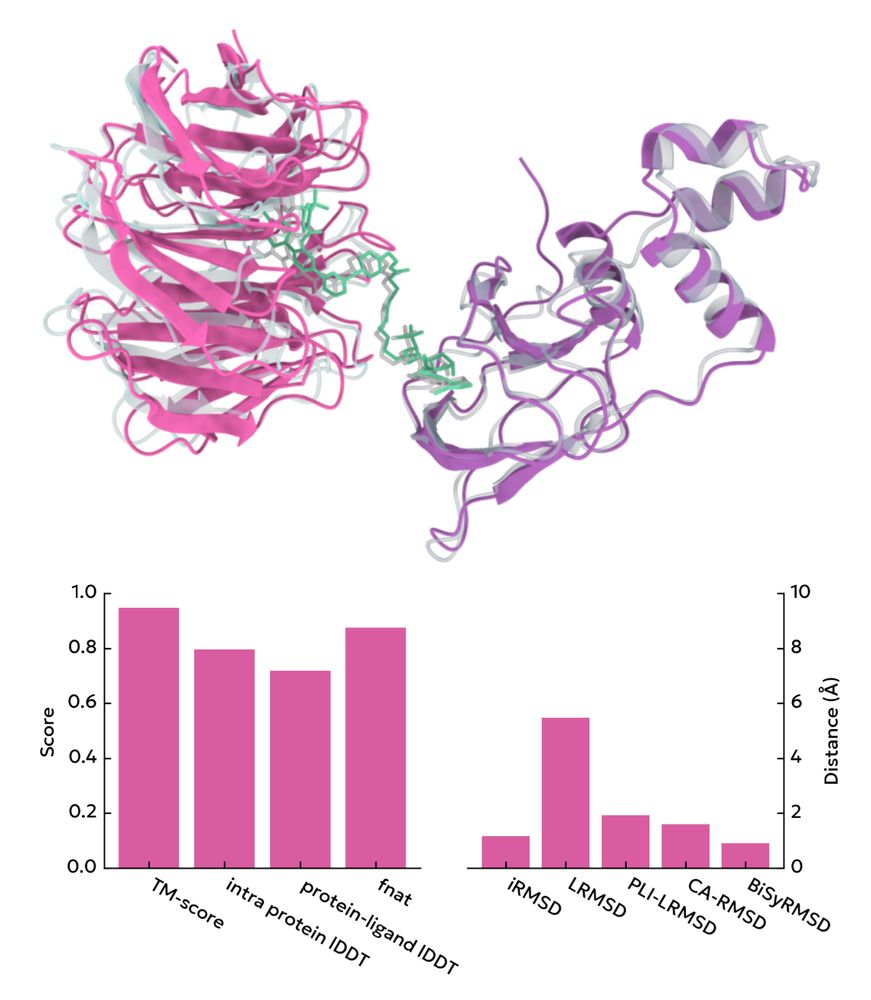
Screencap from the homepage showing various structural similarity metrics between a pose and a reference structure
A single python package for calculation of quality metrics like DockQ, LDDT, RMSD, & others
peppr.vant.ai
Very true!
03.04.2025 05:28 — 👍 2 🔁 0 💬 0 📌 0
Run BioEmu in Colab - just click "Runtime → Run all"! Our notebook uses ColabFold to generate MSAs, BioEmu to predict trajectories, and Foldseek to cluster conformations.
Thanks @jjimenezluna.bsky.social for the help!
🌐 colab.research.google.com/github/sokry...
📄 www.biorxiv.org/content/10.1...
We’re looking for fellow nerds to help us make disordered proteins druggable. Love #IDPs, #MD, #NMR, #AI, #DrugDiscovery, or #MassSpec? Let’s talk.
@bindresearch.org is hiring across the board: bindresearch.org/careers/
#compchem #compbio #NMRChat #TeamMassSpec #chemjobs @chemjobber.bsky.social
Structural bases for Nuclear Factor 1-X activation and DNA recognition. Prototypic insight into the NFI transcription factor family https://www.biorxiv.org/content/10.1101/2025.03.21.644642v1
22.03.2025 03:45 — 👍 1 🔁 3 💬 0 📌 0Structural bases for Nuclear Factor 1-X activation and DNA recognition. Prototypic insight into the NFI transcription factor family https://www.biorxiv.org/content/10.1101/2025.03.21.644642v1
22.03.2025 03:45 — 👍 2 🔁 1 💬 0 📌 0I missed this when it came out. (Or I’ve forgotten it. Hard to know which… 😄)
Either way, perhaps the future is more research alliances with Europe? (And away from the US.)
NZ is associated to pillar 2 of the Horizon programme: research-and-innovation.ec.europa.eu/strategy/str...
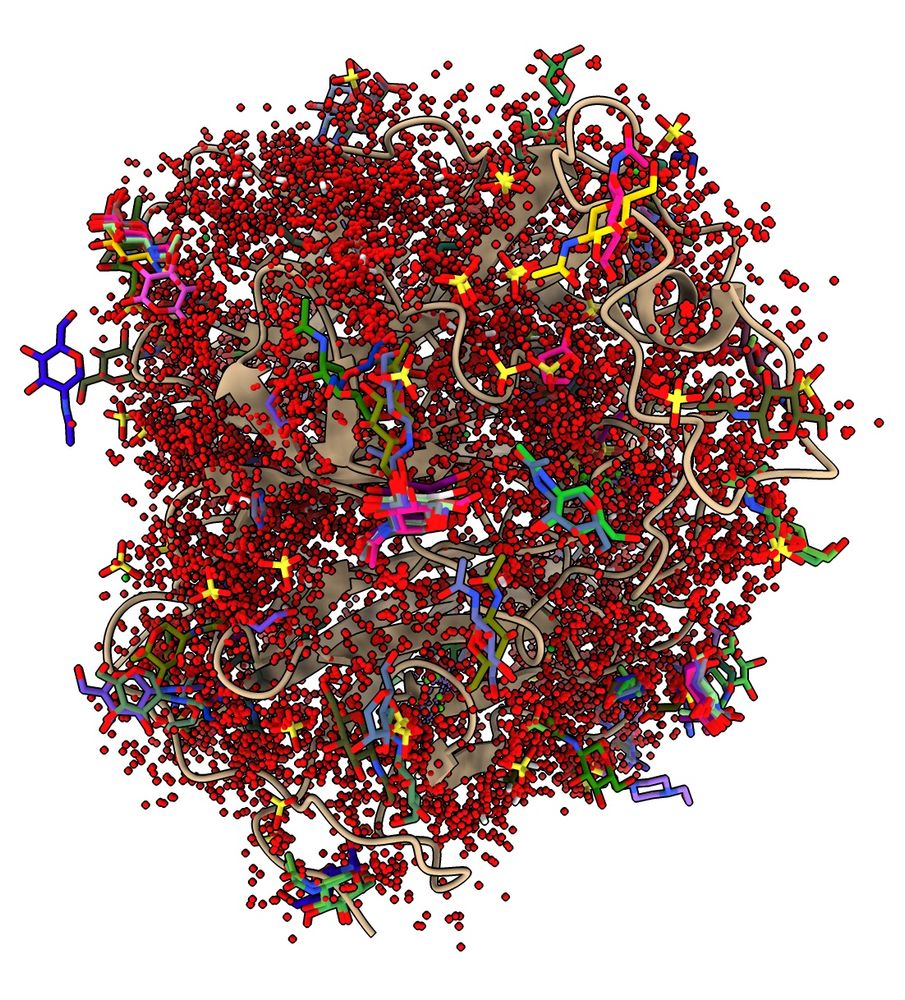
Nipah virus G protein with 16813 ligands (mostly waters) mapped from similar structures.
ChimeraX can run Foldseek to find similar structures, such as distantly related homologs, and analyze the results, for example, mapping all ligands onto your query structure. Here are ligands mapped onto Nipah virus G protein. www.rbvi.ucsf.edu/chimerax/dat...
06.03.2025 23:06 — 👍 96 🔁 36 💬 2 📌 5
The paper describing our community effort to collect and organize #plumed tutorials has been published in the Journal of Chemical Physics, as part of the Michele Parrinello Festschrift! doi.org/10.1063/5.02...
04.03.2025 14:13 — 👍 31 🔁 11 💬 0 📌 1
this is now in @elife.bsky.social doi.org/10.7554/eLif..., a very good experience and I hope other journals will follow this direction
04.03.2025 09:02 — 👍 9 🔁 2 💬 0 📌 0In this world of shrinking research budgets, academic layoffs and failing universities, Elsevier is still making an obscene profit.
If anything has radicalised me to green it's these numbers.
www.researchprofessionalnews.com/rr-news-worl...

It is with sadness I share that my long-term collaborator and friend prof. Per Jemth passed away last week after a short time of illness. He is greatly missed.
19.02.2025 07:44 — 👍 13 🔁 4 💬 5 📌 3We are building high-throughput tools and public datasets to make disordered proteins druggable for everyone. Want to drug the undruggable with us? We’d love to hear from you! Reach out at bindresearch.org.
11.02.2025 09:24 — 👍 7 🔁 1 💬 0 📌 0