
Achira is growing! We’re looking for talented software engineers, ML research engineers, and AI/ML scientists to join our team in building foundation simulation models to power the future of drug discovery.
Apply at achira.ai
@jessbwhite.bsky.social
Current: Tri-I CBM student in @jchodera@bsky.social and Wesley Tansey labs Previous: SWTX | OrbiMed | WeillCornellGS | BofAML | Penn | Andover Employer-mandated disclaimer: choderalab.org/disclaimer

Achira is growing! We’re looking for talented software engineers, ML research engineers, and AI/ML scientists to join our team in building foundation simulation models to power the future of drug discovery.
Apply at achira.ai

How many crystal structures do you need to trust your docking results?

Figure 2

Figure 3

Figure 4
How many crystal structures do you need to trust your docking results? [new]
Accurate drug docking needs crystal struct. Template docking via mol. similarity helps w/ ltd. data.


@jengreitz.bsky.social l & my lab want to co-hire a computational biologist/biostatistician with project management expertise to help map the regulatory code of the human genome and discover genetic mechanisms of disease.
Details below
careersearch.stanford.edu/jobs/computa...
Plz RT
📌
17.08.2025 00:52 — 👍 0 🔁 0 💬 0 📌 0pls RT for reach! can’t wait to see y’all there 😁
@acs.org #CompChem
📌
04.07.2025 09:30 — 👍 0 🔁 0 💬 0 📌 0📌
01.07.2025 16:43 — 👍 0 🔁 0 💬 0 📌 0📌
18.06.2025 13:45 — 👍 1 🔁 0 💬 0 📌 0📌
15.06.2025 05:25 — 👍 0 🔁 0 💬 0 📌 0📌
11.06.2025 12:54 — 👍 0 🔁 0 💬 0 📌 0The steady deterioration of First Amendment rights and the conflation of violence with dissent is a result not just of an authoritarian regime but of a press culture that is feckless at best and authoritarian-fluffers at worst. The New York Times is beyond redemption.
23.05.2025 16:34 — 👍 3187 🔁 746 💬 63 📌 25Grateful for the opportunity to present my work to such an engaged audience! Find out more here: bit.ly/4kcOQnX #Kinase2025
13.05.2025 17:30 — 👍 10 🔁 0 💬 0 📌 0📌
20.04.2025 10:36 — 👍 0 🔁 0 💬 0 📌 0This could be incredibly dangerous
11.04.2025 02:20 — 👍 1 🔁 0 💬 0 📌 0📌
05.04.2025 00:43 — 👍 3 🔁 0 💬 0 📌 0Communicate your opposition to the cancelation of billions of dollars in pandemic preparedness grants to all of your elected representatives simultaneously here: resist.bot/petitions/PX...
27.03.2025 18:15 — 👍 1 🔁 0 💬 0 📌 0In honor of #MarchMadness, let's talk about how all 8 men's teams playing in Providence this week are threatened by the Musk/Trump Admin.
If you love college sports, you're going to have to stand up for colleges!!!!
This work is now published in J Phys Chem B! Check out our work showing that simulations predict the impact of distal mutations on kinase-inhibitor binding, and our experimental NanoBRET dataset of 94 kinase mutations that provide a benchmark for future methods. Link:
pubs.acs.org/doi/full/10....

Musk Said No One Has Died Since Aid Was Cut. That Isn’t True. www.nytimes.com/interactive/...
15.03.2025 16:44 — 👍 0 🔁 0 💬 0 📌 0
Francis Collins, the NIH Director for 12 years, led the Human Genome Project and other NIH efforts for 32 years, resigned today. Key words from his resignation letter
www.nytimes.com/2025/03/01/u...
📌
22.02.2025 17:49 — 👍 0 🔁 0 💬 0 📌 0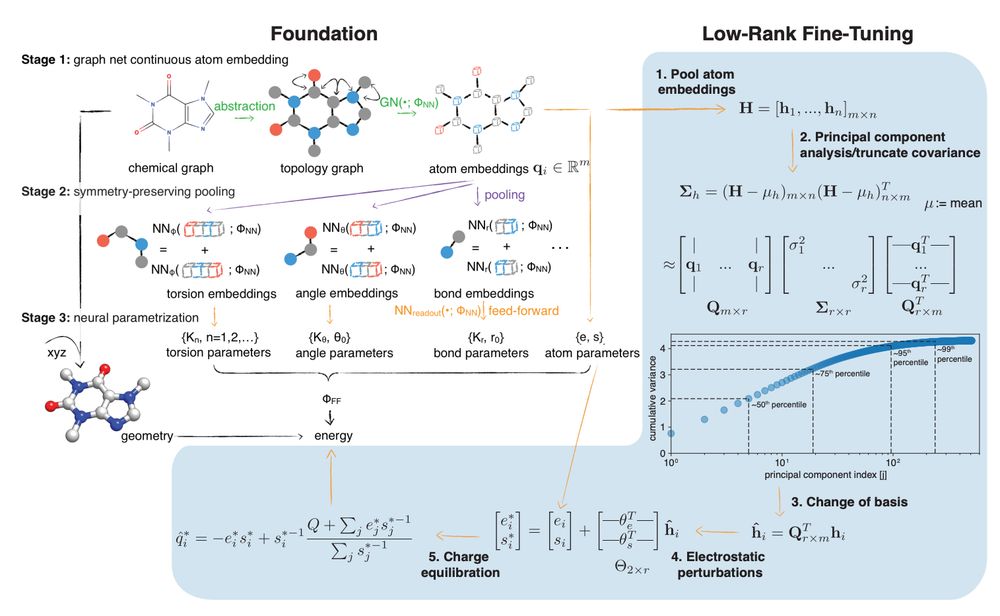
Figure 1 from arXiv preprint https://doi.org/10.1101/2025.01.06.631610 Fig. 1 Espaloma is an end-to-end differentiable molecular mechanics parameter assignment scheme for arbitrary organic molecules. Espaloma (extensible surrogate potential optimized by message-passing) is a modular approach for directly computing molecular mechanics force field parameters FFF from a chemical graph G such as a small molecule or biopolymer via a process that is fully differentiable in the model parameters FNN. In Stage 1, a graph neural network is used to generate continuous latent atom embeddings describing local chemical environments from the chemical graph. In Stage 2, these atom embeddings are transformed into feature vectors that preserve appropriate symmetries for atom, bond, angle, and proper/improper torsion inference via Janossy pooling.54 In Stage 3, molecular mechanics parameters are directly predicted from these feature vectors using feed-forward neural networks. This parameter assignment process is performed once per molecular species, allowing the potential energy to be rapidly computed using standard molecular mechanics or molecular dynamics frameworks thereafter. The collection of parameters FNN describing the espaloma model can be considered as the equivalent complete specification of a traditional molecular mechanics force field such as GAFF38,39/AM1-BCC55,56 in that it encodes the equivalent of traditional typing rules, parameter assignment tables, and even partial charge models. Reproduced from ref. 49 with permission from the Royal Society of Chemistry.
Everything is chaos, but I wanted to share some awesome recent science from the lab that hints at where the future of biomolecular simulation is headed:
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
🧵 on our preprint: Zhang & Skolnick's TM score for comparing model of protein to experimental structures of same protein. The d_j are essentially the same as the aligned error in Alphafold. After any structure alignment, it's the displacement of model Calpha from experimental Calpha of residue j.
16.02.2025 02:13 — 👍 60 🔁 16 💬 4 📌 4Excited to be in LA for #BPS2025 @biophysicalsoc.bsky.social! I’ll be presenting a poster on our high-throughput fluorescence-based assay for kinase inhibitor binding in the West Exhibit Hall on Monday at 2:45 PM (board B60).
16.02.2025 18:47 — 👍 8 🔁 4 💬 1 📌 1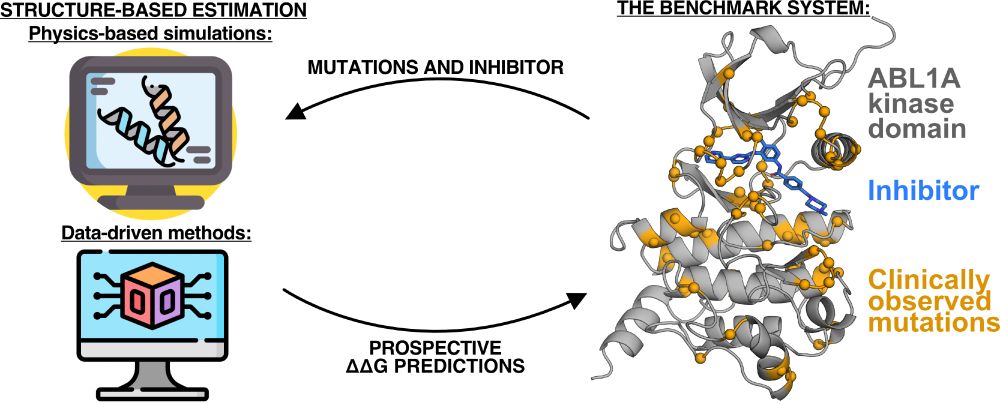
I'll be presenting my work prospectively studying clinical mutations and their impact on kinase inhibitor binding at #bps2025! Come see my talk on Wednesday morning @ 8:45am.
16.02.2025 18:53 — 👍 35 🔁 9 💬 1 📌 01. Today the NIH director issued a new directive slashing overhead rates to 15%.
I want to provide some context on what that means and why it matters.
grants.nih.gov/grants/guide...
#MLCB2025 will be Sept 10-11 at @nygenome.org in NYC! Paper deadline June 1st & in-person registration will open in May. Please sign up for our mailing list groups.google.com/g/mlcb/ for future announcements. More details at mlcb.github.io. Please RP!
27.01.2025 18:40 — 👍 33 🔁 14 💬 0 📌 1📌
20.01.2025 20:26 — 👍 1 🔁 0 💬 0 📌 0
🏁 The antiviral challenge is live! 🏁
Ready to test your skills on new data? Hosted in partnership with @asapdiscovery.bsky.social and @omsf.io, we've prepared detailed notebooks showcasing how to format your data and submit your solutions. 🧑💻
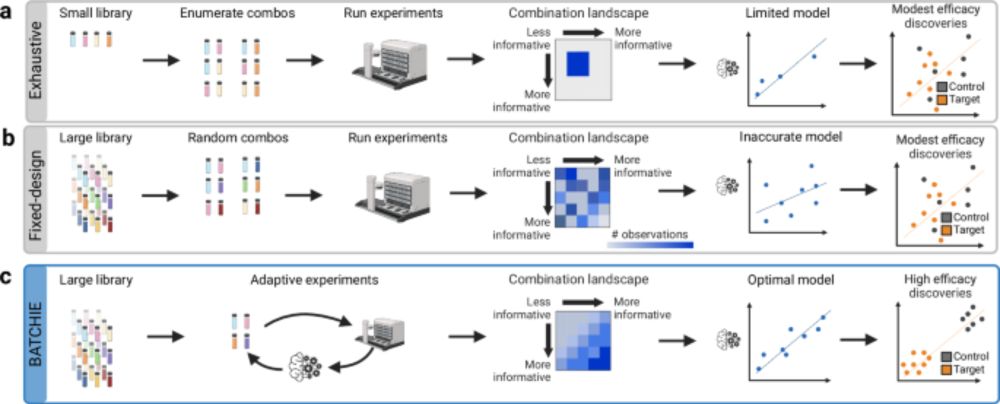
Happy to have been a member of the BATCHIE team who worked on this recent publication featured by @MSKLibrary: A Bayesian active learning platform for scalable combination drug screens dx.doi.org/10.1038/s414...
14.01.2025 15:04 — 👍 11 🔁 1 💬 0 📌 0