Glad to receive a PhD prize from the @gensocuk.bsky.social for the work I lead on the origin and diversification of stratified epithelia.
Thanks to @frelab.bsky.social , the lab, and all collaborators involved.
Special thanks to @candice-merle.bsky.social for her help throughout the project.
26.01.2026 14:31 —
👍 5
🔁 2
💬 0
📌 0
I recently moved to Basel and was awarded an @embo.org Postdoctoral Fellowship to join the lab of Matthias Lutolf at the Institute of Human Biology. I will use data-driven approaches to investigate how micro-environmental cues shape human epithelial organoid maturation.
14.01.2026 15:47 —
👍 3
🔁 0
💬 2
📌 0
Many thanks to @frelab.bsky.social, all collaborators, interns, and researchers involved in these five years!
14.01.2026 15:47 —
👍 1
🔁 0
💬 1
📌 0
1. Excited to share CONCORD, out in @NatureBiotech, an ML framework for single-cell analysis addressing integration, dimensionality reduction, and denoising in one go by @qinzhu1.🔗 www.nature.com/articles/s41... Check out this CONCORD model of worm development resolving differentiation trajectories:
13.01.2026 13:08 —
👍 13
🔁 4
💬 1
📌 0
*New preprint alert* uncovering a mechanical pacemaker that synchronizes nephron formation with branching of the kidney's epithelial tubule tree. Read below to learn about this twisty journey lead by Sam Grindel and Sachin Davis in the lab. [Movie by Nils Lindstrom]
www.biorxiv.org/content/10.1...
13.12.2025 00:08 —
👍 39
🔁 10
💬 2
📌 3
Check out our beautiful story on tumour clonal dynamics with barcodes, performed in the @frelab.bsky.social by my super talented post-doc @candice-merle.bsky.social and @ivanditerlizzi.bsky.social from @rulands.bsky.social lab!
#barcodes #plasticity
03.12.2025 08:55 —
👍 7
🔁 1
💬 0
📌 1

An amazing preprint on molecular evolution (p63-Notch) of Epithelial Multilayering
Cross-organ (14 in🐭), Cross-species (7🪰🐟x2🐸🐥🐭👤) single-cell transcriptomics
p63+Jag2+ basal signal sendor▶️ p63-Hes1+ suprabasal receivers🐭
bioRxiv 2025
www.biorxiv.org/content/10.1...
03.12.2025 12:34 —
👍 9
🔁 3
💬 0
📌 0
Supported by:
@agencerecherche.bsky.social
@frm-officiel.bsky.social
@fondationarc.bsky.social
@cerclefser.bsky.social
@worldwidecancer.bsky.social
@institutcurie.bsky.social
17.11.2025 12:32 —
👍 2
🔁 0
💬 0
📌 0
This work was conducted at @institutcurie.bsky.social
in @frelab.bsky.social. Thank you for supporting my work over the last five years! Big thank also to Candice Merle, the first author of this story, for her help in the experimental work and to our collaborators at @lbmcinlyon.bsky.social.
17.11.2025 12:32 —
👍 0
🔁 0
💬 1
📌 0
Our work unifies decades of organ-specific studies into a single evo-devo framework. It argues for moving beyond organ-centric approaches toward cross-tissue comparisons to reveal conserved mechanisms of epithelial development, homeostasis, and disease.
www.biorxiv.org/content/10.1...
17.11.2025 12:32 —
👍 1
🔁 0
💬 1
📌 0

Taken together, multilayered epithelia development follows a spatial and evolutionary hourglass: basal layers reactivate an ancestral ectodermal program, while suprabasal compartments diversify through modular, lineage-specific innovations.
17.11.2025 12:32 —
👍 1
🔁 0
💬 1
📌 0

Lineage-tracing experiments across all 14 tissues show that activating Notch in basal cells consistently triggers suprabasal commitment, confirming the universality of this regulatory connection in stratified tissues.
17.11.2025 12:32 —
👍 0
🔁 0
💬 1
📌 0
Basal and suprabasal layers are linked by a conserved p63–Notch axis. p63 maintains basal identity and induces Notch ligands, whereas Notch signaling drives suprabasal commitment.
17.11.2025 12:32 —
👍 1
🔁 0
💬 1
📌 0

In contrast, suprabasal compartments show strong enrichment for tissue-specific and architecture-specific transcriptional modules. Functional diversification emerges primarily in suprabasal layers and follow a Russian-doll organization.
17.11.2025 12:32 —
👍 0
🔁 0
💬 1
📌 0
This basal program has been repeatedly redeployed (heterotopy) in endoderm- and mesoderm-derived epithelia, allowing these tissues to initiate multilayering outside their original lineage of origin.
17.11.2025 12:32 —
👍 0
🔁 0
💬 1
📌 0
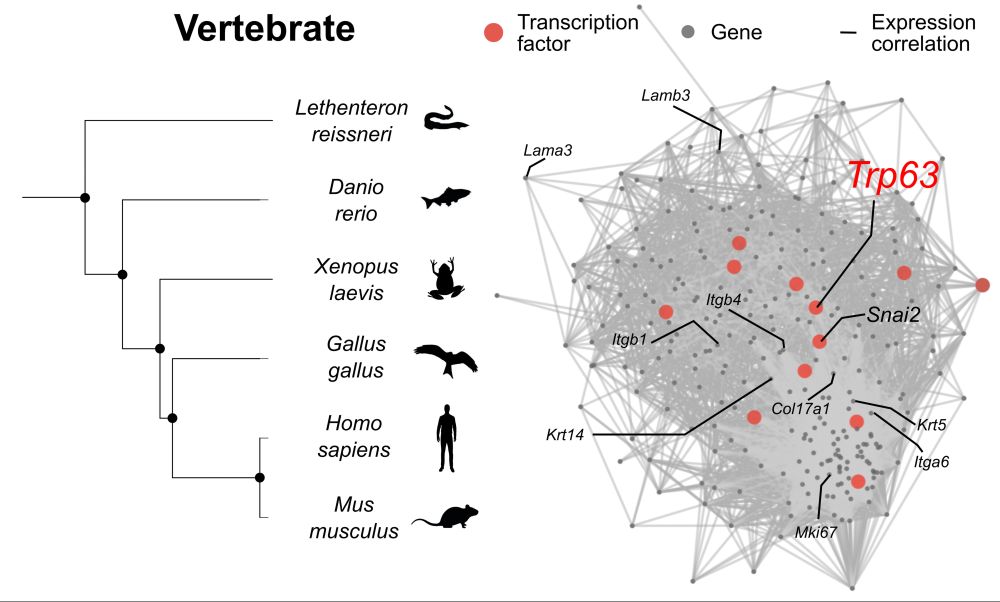
We find that this conserved GRN is evolutionarily compartmentalized. Basal cells consistently deploy an ancestral ectodermal regulatory module centered on p63.
17.11.2025 12:32 —
👍 1
🔁 0
💬 1
📌 0
Comparative analyses across lamprey, zebrafish, xenopus, chicken, mouse, and human reveal that multilayered epithelia rely on a deeply conserved set of genes. This suggests that the molecular foundations of epithelial stratification were established early in vertebrate evolution.
17.11.2025 12:32 —
👍 1
🔁 0
💬 1
📌 0

Multilayered epithelia emerge from ectoderm, endoderm, and mesoderm, yet all adopt the same basic architecture: a basal compartment supporting one or more differentiated suprabasal layers. Do these different organs use distinct mechanisms, or a shared regulatory program?
17.11.2025 12:32 —
👍 1
🔁 0
💬 1
📌 0
🚨 New preprint!
We built a single-cell atlas of 14 multilayered epithelia and revealed a conserved transcriptomic program guiding tissue architecture and fate composition. Our work brings decades of tissue-specific studies together into a unified evo-devo framework.
www.biorxiv.org/content/10.1...
17.11.2025 12:32 —
👍 41
🔁 12
💬 1
📌 0
Supported by:
@agencerecherche.bsky.social
@frm-officiel.bsky.social
@fondationarc.bsky.social
@cerclefser.bsky.social
@worldwidecancer.bsky.social
@institutcurie.bsky.social
16.11.2025 09:21 —
👍 0
🔁 0
💬 0
📌 0

Our paper is on the cover of @cp-devcell.bsky.social . Image: embryonic murine salivary-gland explants stained for fate determinants; p63 (cyan) and HES1 (yellow). Thanks to everyone involved.
doi.org/10.1016/j.de...
06.10.2025 19:38 —
👍 52
🔁 10
💬 2
📌 0

"Contractile fibroblasts form a transient niche for the branching mammary epithelium."
Now out: rdcu.be/eIIKD
A great contribution from Jakub Sumbal, showing how stromal cells, and diverse fibroblast subsets, regulate branching morphogenesis.
@sumbalovakoledova.bsky.social @frelab.bsky.social
30.09.2025 06:13 —
👍 3
🔁 0
💬 0
📌 0
From a FACS-contaminating cell population to a multi-organ project — learn the Behind the Story of our work on conserved signals in epithelial organogenesis 👉 www.sciencedirect.com/science/arti...
08.09.2025 06:23 —
👍 2
🔁 0
💬 0
📌 0
Yes! We think this mechanism goes way beyond glands. We're now exploring a broader range of tissues, stay tuned for what's coming next!
04.07.2025 06:16 —
👍 1
🔁 0
💬 0
📌 0
Great question! We haven’t inferred or directly measured mechanical forces yet, but in a follow-up project we’re actively investigating the mechanical component of the symmetry-breaking event. Very curious to see how it ties into YAP localization and happy to discuss more!
04.07.2025 06:10 —
👍 1
🔁 0
💬 0
📌 0

Students (Master's/PhD), do not miss this fantastic Developmental Biology course of @sorbonne-universite.fr and @institutcurie.bsky.social - It is FREE and open to international students, however, room and board + travel are not included.
training.institut-curie.org/courses/deve...
13.06.2025 16:55 —
👍 69
🔁 49
💬 2
📌 7
This work was funded by @institutcurie.bsky.social, @cnrsbiologie.bsky.social, @psl-univ.bsky.social, @agencerecherche.bsky.social , @frm-officiel.bsky.social, @cerclefser.bsky.social, @fondationarc.bsky.social and @worldwidecancer.bsky.social.
01.07.2025 06:36 —
👍 3
🔁 0
💬 0
📌 0
















