
New preprint: The first whole-body map of both DNA methylation and 3D genome organization at single-cell resolution across 16 human tissues, with in-depth analyses of cell type diversity of these epigenome modalities at unprecedented resolutions. (1/5)
25.03.2025 15:16 —
👍 6
🔁 4
💬 1
📌 0
Thanks for the support from @arcinstitute.org. Especially want to thank the people not on the author list but helped from all the aspects - Rajesh Ilango, Scott Newins, @nick-youngblut.bsky.social, Dave Burke, @brianplosky.bsky.social, Joe Caputo, @ruochiz.bsky.social, and @siyuhe.bsky.social.
25.03.2025 15:49 —
👍 1
🔁 0
💬 0
📌 0
10/10 Thankful to the great team, otherwise the large effort would never be achieved. This work is led together with my teammates in Joe Ecker‘s lab and @jesserdixon.bsky.social lab. The data was generated back during my PhD and analysis and manuscript done after starting at Arc.
25.03.2025 15:49 —
👍 1
🔁 0
💬 1
📌 0
Human tissue browser
8/10 To facilitate the widespread use of this resource, we have developed a web browser for readers to explore the two data modalities across different tissues, major cell types, and cell subtypes (humancellepigenomeatlas.arcinstitute.org)
25.03.2025 15:49 —
👍 0
🔁 0
💬 1
📌 0

7/10 3D genome structures were analyzed in a few dozens of bulk tissues. Here we identified >600k differential loop pixels across major types and subtypes, representing the cell type specific 3D genome architecture at the highest cellular resolution so far.
25.03.2025 15:49 —
👍 1
🔁 0
💬 1
📌 0

6/10 non-CG methylation (mCH) was primarily detected in brian and stem cells. Here we reported mCH across almost all major cell types and show cell type specific signatures at genes and regulatory elements.
25.03.2025 15:49 —
👍 1
🔁 0
💬 1
📌 0
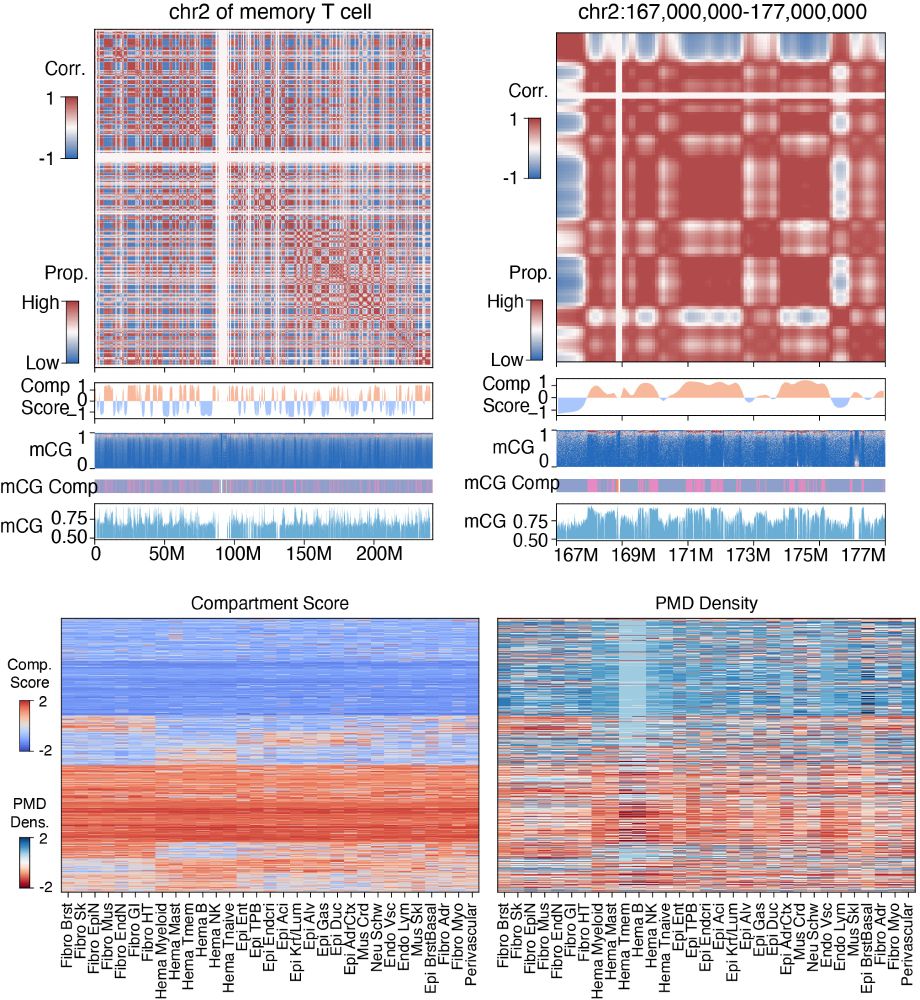
5/10 Partially methylated domains (PMD) were identified in cancer cell lines and placenta. Here we reported PMD-like structures in almost all cell types and generalized it to methylation compartment, which correlated with 3D genome compartment but at a higher genome resolution.
25.03.2025 15:49 —
👍 1
🔁 0
💬 1
📌 0

4/10 The cell identities could be driven by only one modality, or the two modalities can show different cellular identity signatures in the same cell at the same gene. Links between chromatin state and cell fate may not be determined by a single set of parameters.
25.03.2025 15:49 —
👍 1
🔁 0
💬 1
📌 0

3/10 Different Epigenome modalities are considered highly correlated with each other. Here we found that extensive discrepancies can exist between methylation and 3D genome organization in cell type classification.
25.03.2025 15:49 —
👍 1
🔁 0
💬 1
📌 0

2/10 snm3C-seq on 16 organs generate 86,689 chromatin conformation and DNA methylome joint profiles. Integration with 24 published scRNA datasets annotate 206 cell subtypes. These produced the whole genome methylation and 3D structure at unprecedented cell-type resolution.
25.03.2025 15:49 —
👍 1
🔁 0
💬 1
📌 0
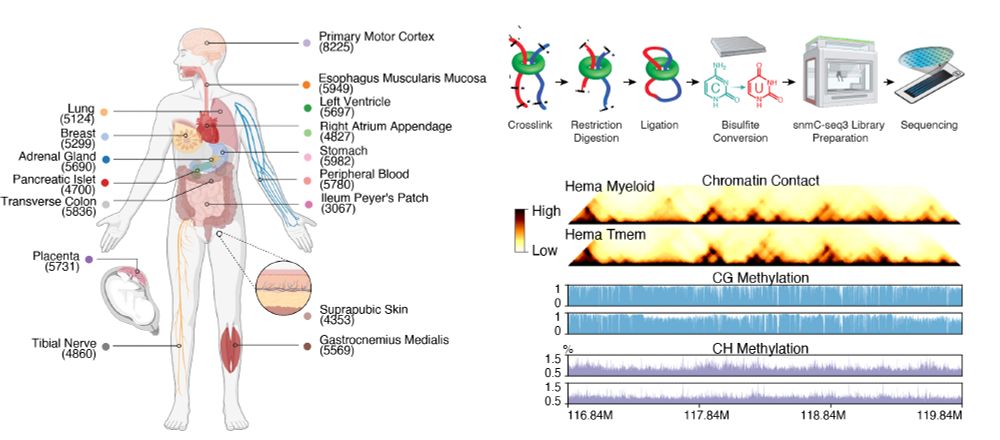
1/10 Excited to share our latest - the first whole-body map of both DNA methylation and 3D genome at single-cell resolution.
25.03.2025 15:49 —
👍 8
🔁 5
💬 1
📌 1








