We had a few lightening talks presenting the community initiatives supported by OMSF: Free Energy Workshop (feworkshop.org) by Jonah Vilseck, BPDMC (bpdmc.org) by Chris Ball, @bitsinbio.bsky.social by Nelly Tian, and OpenRosetta by Julia Koehler Leman.
09.05.2025 20:45 — 👍 1 🔁 0 💬 0 📌 0

After our last coffee break, @dwhswenson.net is updating everyone about progress we've made on our #NSF POSE grant, including the release of the first batch of playbooks available at playbooks.omsf.io. We started with the contributor, developer and benchmarking playbooks.
09.05.2025 19:54 — 👍 2 🔁 0 💬 1 📌 0

Woody Sherman from Psivant provided an industry perspective on molecular modelling, open science, @openfold.io consortium, @omsf.io and future directions for computational molecular sciences. We also got richer for a new expression that received hearty approval -- "free energy landscape artist"
09.05.2025 19:45 — 👍 1 🔁 0 💬 1 📌 0
Aniket Margakar from Boehringer demonstrated how they use @omsf.io tools in house, praising both the tools and teams -- that's what we like to hear! He highly recommends NAGL and bespokefit for fast charge assignment and parameterization, and @openfree.energy for FE calcs
09.05.2025 18:51 — 👍 1 🔁 1 💬 1 📌 1
@jenkescheen.bsky.social showcased his work on @asapdiscovery.bsky.social and more recently with CharmTx, combining ML and more traditional free energy calculations. A lot, if not all, of his work is based on @openfree.energy, #openforcefield, and #alchemsicale. Read his papers for more details!
09.05.2025 18:46 — 👍 1 🔁 2 💬 1 📌 0

GitHub - openforcefield/joint-demo: Materials for the joint demo at the 2025 OMSF Symposium
Materials for the joint demo at the 2025 OMSF Symposium - openforcefield/joint-demo
After refueling with some sandwiches, Matt Thompson ran a demo connecting OMSF tools in a working workflow (!). You can find the demo notebook here and try it yourself: github.com/openforcefie.... Feedback welcome!
09.05.2025 18:33 — 👍 1 🔁 1 💬 1 📌 0

@moalquraishi.bsky.social shared some challenges encountered and learnings gained through painful data preparation and multiple training experiments for the next release of OpenFold, which is coming soon!
09.05.2025 16:17 — 👍 1 🔁 0 💬 1 📌 0

Ryan Renslow and Hugo MacDermott-Opeskin gave a gentle intro to our latest project @openadmet.bsky.social in collaboration with QBI UCSF and Octant Bio, funded by ARPA-H.
09.05.2025 16:09 — 👍 2 🔁 0 💬 1 📌 0
we are having a blast! keynotes from @openfree.energy , @openfold.io , and @openadmet.bsky.social, and Open Force Field have been stellar. @lilyminium.bsky.social and @ialibay.bsky.social - we are so proud!
09.05.2025 15:41 — 👍 7 🔁 2 💬 0 📌 0


Next in line is @ialibay.bsky.social with an overview of Open Free Energy and the latest plans and developments. Did I say we have a full house in the lovely Museum of Science in grey Boston?
09.05.2025 13:57 — 👍 3 🔁 0 💬 1 📌 0


In short, the new release is coming soon, the team is working hard on consistent force fields for small molecules and (bio)polymers, plus some new tools in the belt, like #pablo.
09.05.2025 13:54 — 👍 1 🔁 0 💬 1 📌 0
@omsf.io 3rd annual symposium kicks off with @michaelshirts.bsky.social and @lilyminium.bsky.social with the latest update on #OpenFF
09.05.2025 13:15 — 👍 6 🔁 1 💬 1 📌 0

On my way to Boston for the annual in-person @omsf.io catch-up! I'm looking forward to seeing everyone and hearing what everyone has been up to, there'll be lots to share!
07.05.2025 20:55 — 👍 1 🔁 0 💬 0 📌 0
And fantastic to hear from @karmencj.bsky.social on the experiences and challenges of building research infrastructure with @omsf.io !
07.04.2025 19:51 — 👍 1 🔁 1 💬 0 📌 0
What is a better first post on the platform than inviting folks to join us at our third annual symposium again in Boston! There's a lot happening at @omsf.io -- spend a day with us and find out all about it. There will be cookies too!
05.02.2025 21:41 — 👍 3 🔁 1 💬 0 📌 0
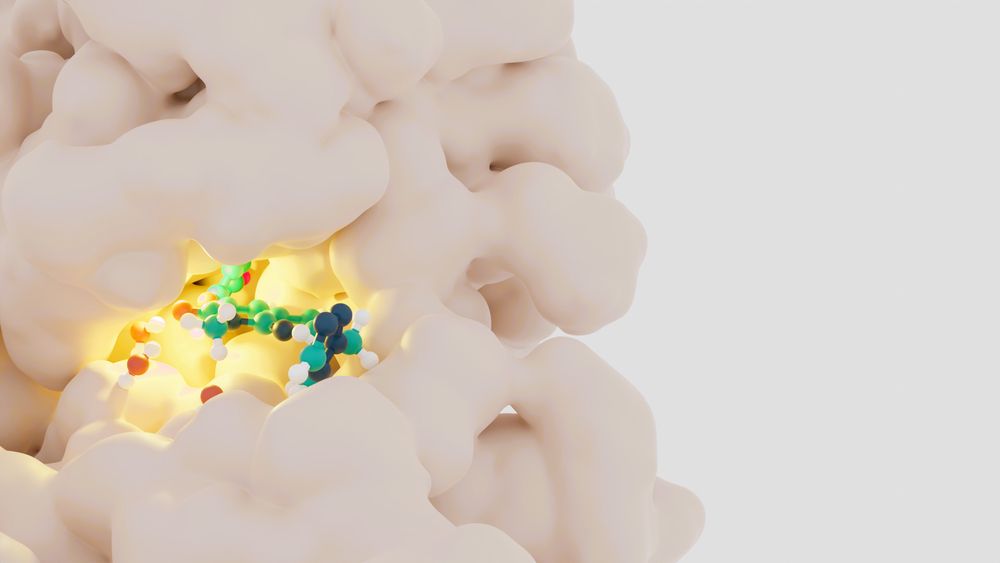
Artistic rendering of a biochemical model: a small molecule ligand, shown as a ball-and-stick model colored by element, is bound in a pocket in a protein surface, shown as a space filling model colored off-white.
We're changing the field of #compchem by creating free and open-source software for performing alchemical free energy calculations. Our flagship protocol calculates relative binding free energies of protein-ligand systems. Try it out in your browser: colab.research.google.com/github/OpenF...
04.02.2025 14:46 — 👍 30 🔁 14 💬 0 📌 0
We are now on Bluesky! OMSF supports the ever-growing ecosystem of open source molecular software. We love all things #opensource, #science, and #software - if you do too, follow us!
28.09.2023 19:27 — 👍 17 🔁 6 💬 0 📌 2
I use robots and biophysics to study cancer therapies in the Chodera Lab at Memorial Sloan Kettering Cancer Center. Views are my own.
Founder of Datalayer https://datalayer.ai
Not-for-profit Focused Research Organisation (FRO) turning disordered proteins into viable drug targets | London, UK | https://bindresearch.org/
The funder-researcher collaboration and open-access publisher for research in the life and biomedical sciences.
Follow @eLifeCommunity.bsky.social
Computational chemist interested in designing and using tools for structure based drug design. Free energies are cool
Computational chemist/structural bioinformatician working on improving molecular simulation at MRC Laboratory of Molecular Biology. jgreener64.github.io
Prof at UC San Diego's Skaggs School of Pharmacy and Pharmaceutical Sciences.
Concepts, tools and methods: stat thermo of binding. GIST. BindingDB. Open Force Field. Other stuff...
Does some ceramics, i.e., potters around.
Professor @pittchem.bsky.social. Computational biophysicist, leading @westpasoftware.bsky.social development for weighted ensemble rare-event sampling, Amber force field developer.
Computational chemistry and biophysics. Empirical force fields, CADD, RNA, carbohydrates, and methods developments including SILCS (site identification by ligand competitive saturation, See SilcsBio LLC) and enhanced-sampling GCMC. And I surf....
Computational biophysicist, prof at KTH, Stockholm. Loves membrane proteins and mathematical models. Mom of twin toddlers. 🇫🇷🇸🇪
Prof. of Biophysics. KTH, Linköping & Stockholm U. Chair IVA Chapter VII. Director National Academic Infrastructure for Supercomputing in Sweden. AI, HPC & Life Sciences.
Theoretical Chemistry research group @lct-umr7616.bsky.social, Sorbonne Université & CNRS| Led by Prof. Piquemal (@jppiquem.bsky.social)|
#compchem #HPC #MachineLearning #quantumcomputing
Website: https://piquemalresearch.com
Head of Computational Structural Biology Lab @pasteur.fr. ERC-CoG 2022. Research Director DR2 @cnrs.bsky.social. Founder/developer @plumed.org
Computational chemist l PhD from John Chodera's group @ MSKCC
Education @ Schrödinger | Comp Med Chem School Co-Founder and Organizer (compmedchem.org)
UCSF ChimeraX - software for visualizing biomolecules
Scientist. Frog hunter. Founder and Chief Executive of Structural Genomics Consortium.
A public-private partnership seeking to accelerate drug discovery through pre-competitive, open access research. More about us at thesgc.org
Computational biophysicists at Arizona State University; MDAnalysis (@MDAnalysis.bsky.social) developer. Opinions my own.
Research Group: https://becksteinlab.physics.asu.edu/
MDAnalysis: https://www.mdanalysis.org/












