
Who lived in a pineapple under the sea?
All of our ancestors
www.science.org/doi/10.1126/...
@tralee-sci.bsky.social
Post doc in the Ecker lab @ The Salk Institute. PhD in the Bailey-Serres lab @ UCR. Posts include papers I'm reading and things that I 3D print @TrALEE_Sci on Twitter

Who lived in a pineapple under the sea?
All of our ancestors
www.science.org/doi/10.1126/...

We like to introduce map3C, developed by Joseph Galasso, that drastically improves the mapping and contact calling performance of snm3C-seq and now enables accurate 3D genome modeling. map3C was developed in collaboration with
Jason Ernst and Frank Alber. www.biorxiv.org/content/10.1...
New pre-print from the team!
The manuscript is @emma-raven.bsky.social's PhD work showing that whether a leaf is a carbon sink or a carbon source influences how they execute immune responses.
Have a read!
#PlantScience
@johninnescentre.bsky.social

Congrats, Joe Ecker @salkinstitute.bsky.social, to receiving the McClintock Prize! Joe has been a visionary leader of the field of genetics and genomics – not only for plants – for decades
www.salk.edu/news-release...

Coming to you live from #ISPLORE2025JP Fresh preprint from my lab showing that leaves progressively oxygenate and how this is important for their morphogenesis.Thanks to our collaborators from @Fra_LicO2si lab. #plantscience
www.biorxiv.org/content/10.1...

New OA Article: "A single-cell rice atlas integrates multi-species data to reveal cis-regulatory evolution" rdcu.be/eHce3
Chromatin accessibility in rice & related grasses: how regulatory DNA elements evolve across cell types & species; identifying potential silencers.

A timely article as several recent conference discussions have led to similar thoughts
www.nature.com/articles/s41...
Thank you Rashmi!
01.09.2025 21:48 — 👍 0 🔁 0 💬 0 📌 0
Our #research on #drought #recovery, now published with @springernature.com in @natcomms.nature.com:
Drought recovery in plants triggers a cell-state-specific immune activation.
doi.org/10.1038/s414...
Read thread below 👇

Our new Nature Plants paper is out (and we’re on the cover 😁)!👉 bit.ly/4lS8sOB
By combining scRNA-seq with conserved TF binding (multiDAP) we define gene regulatory networks for 65 cell types across 4 tissues in a wide range of flowering plants!

Thrilled to share our latest work: how plants control growth through activation of a surface-specific growth programme. Thanks @zoenv.bsky.social, @nathan-german.bsky.social and the other coauthors for all the hard work! www.biorxiv.org/content/10.1...
27.08.2025 18:07 — 👍 78 🔁 28 💬 3 📌 1
Happy to see our latest #plantscience review by @aidamaric.bsky.social, @advaitagashe.bsky.social and Johanna onlinr! We describe how epigenetic mechanisms control ethylene signal generation and progression, and how ethylene in turn modulates chromatin.
www.sciencedirect.com/science/arti...
#Kpopdemonhuntersquad
22.08.2025 06:20 — 👍 1 🔁 0 💬 1 📌 0
Plz share! (I am a co-author from NAASC) “An unwelcoming climate & culture at scientific conferences is an obstacle to retaining scientists w/marginalized identities. Here we describe..a professional plant science societies..collaboration to make conferences more inclusive.”
doi.org/10.7554/eLif...
Thanks for the enthusiasm in using our online web browser! Due to the high traffic, we have allocated additional resources for user access, so if anyone encountered a bug with accessing the data, please give it a try again and reach out if additional errors are encountered!
20.08.2025 18:25 — 👍 1 🔁 0 💬 0 📌 0It may arrive sooner than you think, the resources and equipment are there!
20.08.2025 06:49 — 👍 0 🔁 0 💬 0 📌 0a little secret- the actual amount of data produced and filtered using the Drosophila cell atlas standards was >800k but we more stringently filtered it to ~430k for all downstream analyses. We plan to make this additional 400k data availability in NCBI GEO and here arabidopsisdevatlas.salk.edu
20.08.2025 00:55 — 👍 6 🔁 4 💬 0 📌 0Tremendous work by the extremely talented postdocs @TrALEE_Sci @NatanellaE @nobolly .Thank you for putting in a major effort on this “side project” to develop a powerful community resource arabidopsisdevatlas.salk.edu
20.08.2025 00:42 — 👍 8 🔁 3 💬 0 📌 0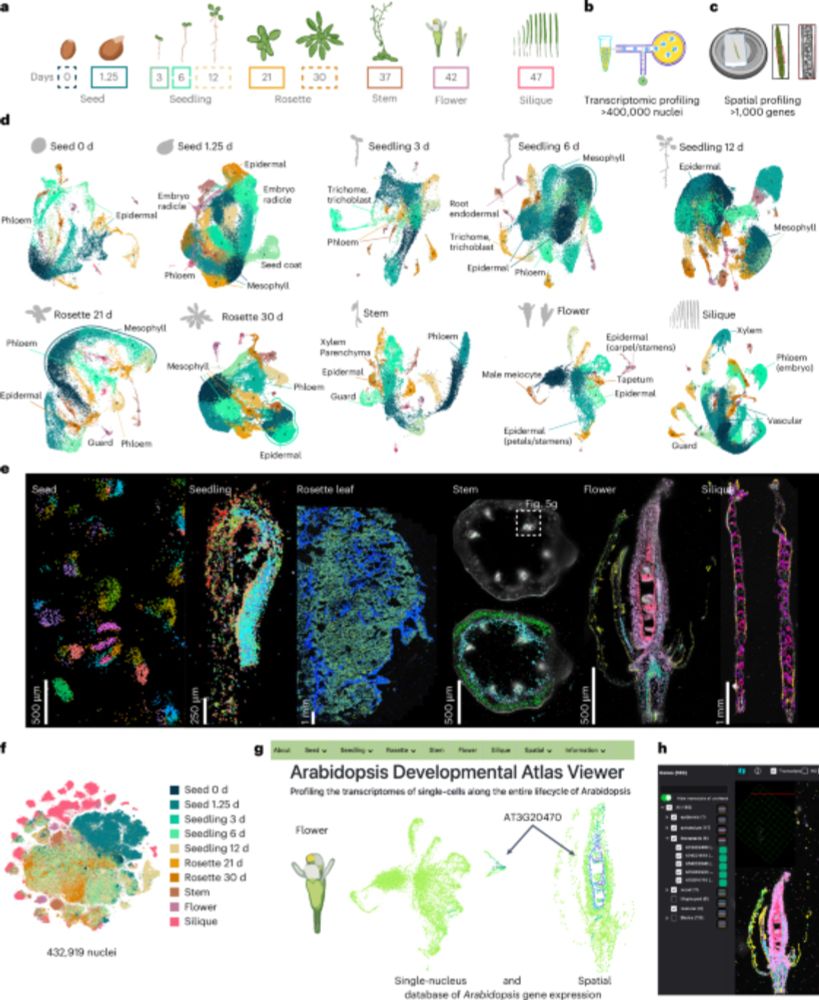
From a gene’s single cell expression–through spatial localization–to novel function, and beyond!
Now out @natplants.nature.com
We built a comprehensive spatial-transcriptomic atlas of Arabidopsis, revealing cell-type identities across organs in unprecedented detail
www.nature.com/articles/s41...
All datasets should be back online as we experienced a surge of traffic, thank you for your patience!
19.08.2025 18:08 — 👍 0 🔁 0 💬 0 📌 0Thanks Alex!
19.08.2025 16:20 — 👍 1 🔁 0 💬 0 📌 0
New OA Resource: "A single-cell, spatial transcriptomic atlas of the Arabidopsis life cycle" rdcu.be/eBmkU
An extensive single-nucleus and spatial transcriptomic atlas of the Arabidopsis life cycle that represents 10 developmental time points in 6 diverse organs.
Thank you Laurence!
It looks like our server may have been overloaded with activity, I will ensure that all of the starts are up and running shortly. One challenge with our manuscript releasing at 2am local time!
Thanks Sagar!
19.08.2025 15:47 — 👍 0 🔁 0 💬 0 📌 0Thanks Kevin!
19.08.2025 15:46 — 👍 0 🔁 0 💬 0 📌 0Thanks Dan!
19.08.2025 15:45 — 👍 1 🔁 0 💬 0 📌 0Thanks Sjon! Sad that I will miss out on isplore this year!
19.08.2025 09:29 — 👍 1 🔁 0 💬 1 📌 0
image of the team of first authors
We hope that this 'initial draft' of a spatially-resolved cell atlas of the Arabidopsis lifecycle will enable further biological discovery and function as a framework for future focused studies. 6/end
19.08.2025 09:17 — 👍 4 🔁 0 💬 0 📌 0We have made all datasets browsable with our web resource arabidopsisdevatlas.salk.edu that empowers anyone to ask where their favorite gene is expressed at single-cell resolution without the need for computational resources or bioinformatic background. 5/n
19.08.2025 09:17 — 👍 16 🔁 4 💬 1 📌 1
spatial expression of markers within the hypocotyl apical hook
We also took a deep dive investigating cell states within the compact yet complex hypocotyl apical hook where we found that our paired spatial + single-cell datasets were key for identifying novel cellular states at the intersection of development and hormonal regulation. 4/n
19.08.2025 09:17 — 👍 2 🔁 0 💬 1 📌 0