GitHub - quadbio/cell-annotator: Automatically annotate cell types, consistently across samples.
Automatically annotate cell types, consistently across samples. - quadbio/cell-annotator
Happy to share CellAnnotator, a lightweight Python package to query OpenAI models for initial cell type annotations: github.com/quadbio/cell...
Inspired by ideas in www.nature.com/articles/s41... and github.com/VPetukhov/GP..., implemented as an scVerse ecosystem package with docs and tutorials.
28.03.2025 16:53 — 👍 8 🔁 4 💬 1 📌 0
From cell lines to full embryos, drug treatments to genetic perturbations, neuron engineering to virtual organoid screens — odds are there’s something in it for you!
Built on flow matching, CellFlow can help guide your next phenotypic screen: biorxiv.org/content/10.1101/2025.04.11.648220v1
23.04.2025 09:26 — 👍 17 🔁 7 💬 1 📌 1
Our paper benchmarking feature selection for scRNA-seq integration and reference usage is out now www.nature.com/articles/s41...!
Keep reading for more about how we did the study and what we found out 🧵 👇
1/16
18.03.2025 15:40 — 👍 41 🔁 17 💬 6 📌 1

Stress alters neuronal balance in the developing brain
Stress hormones, often prescribed before premature delivery, affect the brain development of the embryo
⚡️Our findings have implications for understanding how prenatal stressors can shape brain function and contribute to mental health disorders later in life. A critical step toward bridging the gap between genetics and environment in brain health.
Check out the press release ⬇️
🧵6/6
21.02.2025 10:18 — 👍 1 🔁 0 💬 0 📌 0
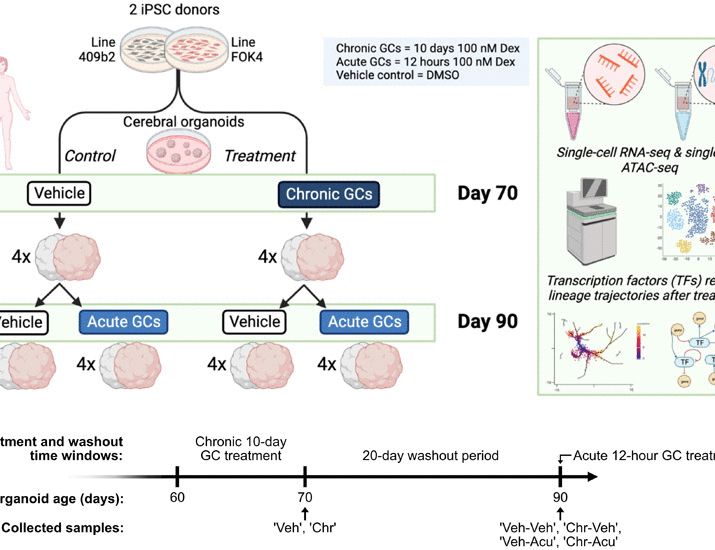
⚠️ Why it matters: We show how environmental factors, like GC exposure, can converge to affect brain development through common molecular mechanisms as genetic risk factors: priming of the inhibitory neuron lineage.
🧵5/6
21.02.2025 10:17 — 👍 1 🔁 0 💬 1 📌 0

We identified PBX3 as an example of a TF that is closely linked to the inhibitory neuron lineage priming. In silico perturbation experiments of multimodal GRNs suggest PBX3 as a potential mediator of the effect. 🧬🔑
🧵4/6
21.02.2025 10:17 — 👍 1 🔁 0 💬 1 📌 0

Our findings show that chronic GC exposure alters lineage specification, with a selective priming of the inhibitory neuron lineage. 🧠
🧵3/6
21.02.2025 10:16 — 👍 1 🔁 0 💬 1 📌 0

In our study, we looked at how chronic GC exposure affects neural differentiation and lineage specification in human neural organoids, using single-cell techniques to map the molecular changes in detail. 🔬🧬
🧵2/6
21.02.2025 10:16 — 👍 1 🔁 0 💬 1 📌 0
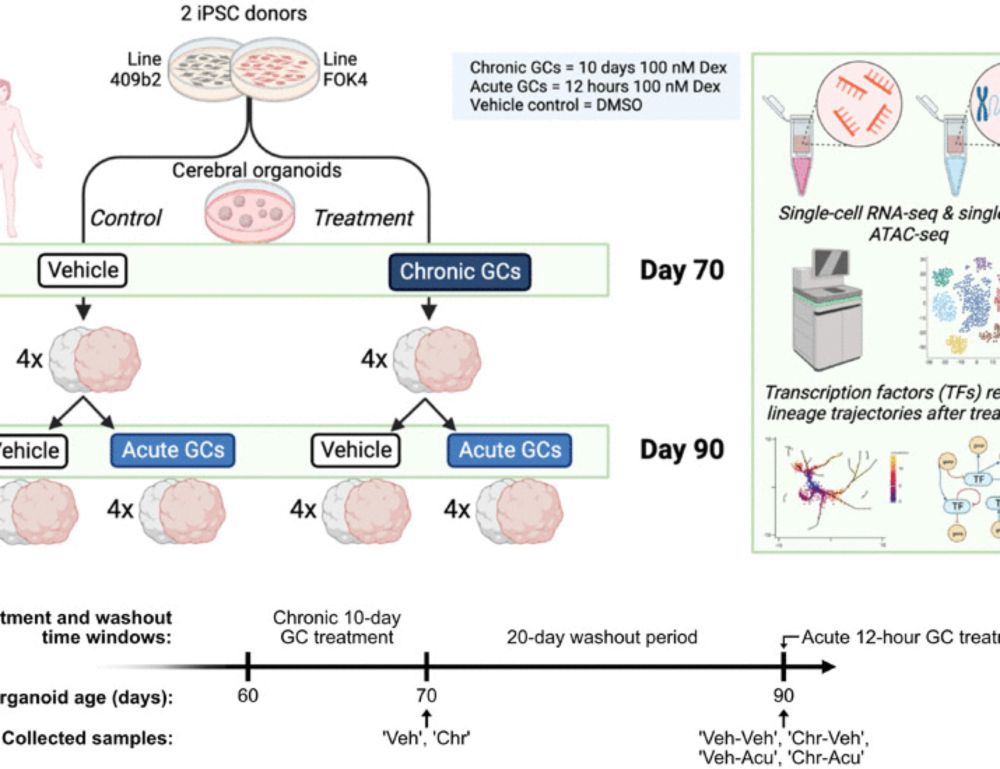
Chronic exposure to glucocorticoids amplifies inhibitory neuron cell fate during human neurodevelopment in organoids
Chronic exposure to glucocorticoids during brain development leads to priming of the inhibitory neuron lineages in organoids.
🚨🧠 New paper alert: Stress alters neuronal balance in the developing brain 🧠🚨
Our latest study in @science.orgAdvances explores how glucocorticoid exposure—a key environmental risk factor—shapes early human brain development using #organoids.
@mpi-psychiatry.bsky.social @drcricru.bsky.social
🧵👇
21.02.2025 10:14 — 👍 14 🔁 5 💬 2 📌 2
Chronic exposure to glucocorticoids amplifies inhibitory neuron cell fate during human neurodevelopment in organoids
Chronic exposure to glucocorticoids during brain development leads to priming of the inhibitory neuron lineages in organoids.
Insightful study by L. Dony @anthikrontira.bsky.social @drcricru.bsky.social @binderlabmpip.bsky.social @silvianeuro.bsky.social @fabiantheis.bsky.social on the effects of chronic exposure to glucocorticoids for neurodevelopment. Organoid model reveals amplification of inhibitory neuron cell fate 🧪
15.02.2025 08:59 — 👍 14 🔁 6 💬 1 📌 0
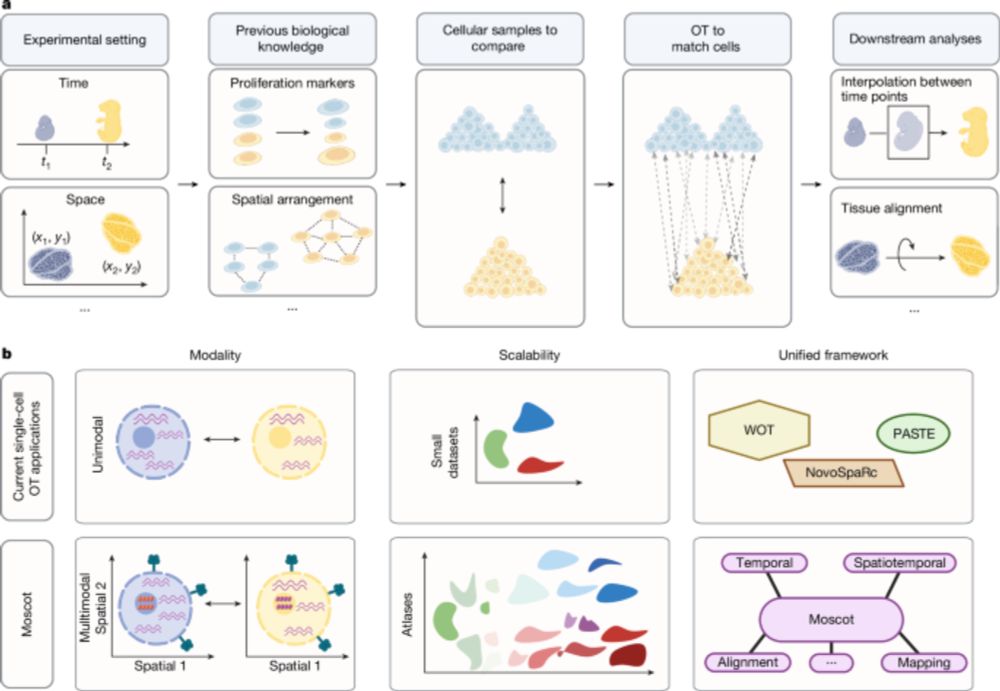
Mapping cells through time and space with moscot - Nature
Moscot is an optimal transport approach that overcomes current limitations of similar methods to enable multimodal, scalable and consistent single-cell analyses of datasets across spatial and temporal...
Good to see moscot-tools.org published in @nature.com ! We made existing Optimal Transport (OT) applications in single-cell genomics scalable and multimodal, added a novel spatiotemporal trajectory inference method and found exciting new biology in the pancreas! tinyurl.com/33zuwsep
23.01.2025 08:41 — 👍 49 🔁 13 💬 1 📌 3

Meinung: »Human Cell Atlas«: Das wird die wichtigste Wissenschaft des 21. Jahrhunderts - Kolumne
Gerade ist ein ganzes Bündel Publikationen aus einem einzigen Forschungsprojekt erschienen. Sie weisen in die Zukunft einer neuen Wissenschaft: Lernende Maschinen helfen jetzt, die Maschinerie des Leb...
If diving into scientific papers isn’t on your pre-Christmas to-do list, @spiegel.de has you covered! 📰 They featured our Neural Organoid Atlas in a piece on the Human Cell Atlas and related advancements in machine learning. 🤖🧬 It’s a great overview of this exciting field and the work behind it.
19.12.2024 16:05 — 👍 10 🔁 7 💬 0 📌 0

The Human Cell Atlas: towards a first draft atlas
In a collection of research articles and related content, the Human Cell Atlas consortium presents tools, data and ideas towards the generation of their first draft atlas of cells in the human body.
And of course to our amazing PIs @graycamplab.bsky.social, @fabiantheis.bsky.social, and Barbara Treutlein for all their guidance and support! 🚀
This work is part of the Human Cell Atlas effort to map all human cells. Explore more @natureportfolio.bsky.social: www.nature.com/immersive/d4...
🧵8/8
21.11.2024 10:17 — 👍 9 🔁 1 💬 0 📌 0
A huge thank you to all collaborators: @chatgtp.bsky.social , @katelynxli.bsky.social, Irena Slišković, Hsiu-Chuan Lin, Malgorzata Santel, Alexander Atamian, @giorgiaquadrato.bsky.social, Jieran Sun, @sergiuppasca.bsky.social & the Human Cell Atlas Organoid Biological Network. 🙏
🧵7/8
21.11.2024 10:15 — 👍 5 🔁 0 💬 1 📌 0

GitHub - devsystemslab/HNOCA-tools: Toolbox of the Human Neural Organoid Cell Atlas
Toolbox of the Human Neural Organoid Cell Atlas . Contribute to devsystemslab/HNOCA-tools development by creating an account on GitHub.
Contextualising New Data 🗺: The atlas serves as a powerful tool for annotating cell types, comparing protocols, and contextualising your new neural organoid scRNA-seq dataset. We provide a Python package (github.com/devsystemsla...) and interactive web interface (through www.archmap.bio#/).
🧵6/8
21.11.2024 10:14 — 👍 7 🔁 0 💬 1 📌 0

Disease Modeling 😷 : Our atlas can be leveraged as a diverse control cohort to contextualise organoid models of disease, helping identify underlying genes and pathways. We found that comprehensive control data is vital for disentangling disease from regional effects.
🧵5/8
21.11.2024 10:14 — 👍 4 🔁 0 💬 1 📌 0

Quantifying Organoid Fidelity 📊: By estimating the transcriptomic similarity between primary and organoid brain counterparts, we provide a robust framework for assessing protocol variation and fidelity across labs and methods. Cell stress seems to be quite dependent on protocol.
🧵4/8
21.11.2024 10:13 — 👍 4 🔁 0 💬 1 📌 0

Identifying Missing Cell States 🔍: We map neural organoid cell types and states to a developing human brain reference, highlighting gaps in the diversity of brain regions present in current in vitro models. ⚠️While the telencephalon seems well-captured, this is not true for all brain regions.
🧵3/8
21.11.2024 10:13 — 👍 4 🔁 0 💬 1 📌 0

Introducing the Human Neural Organoid Cell Atlas 🗺️: We’ve integrated 36 neural organoid datasets across 26 protocols. This produced a comprehensive, integrated reference atlas, including a hierarchical cell-type annotation, partially derived by mapping to primary human 🧠.
🧵2/8
21.11.2024 10:12 — 👍 4 🔁 0 💬 1 📌 0
Comp scientists who are passionate about developing novel tools powered by AI to accelerate discovery & translation.
@www.helmholtz-munich.de
technician Max Planck Institute of Psychiatry, Munich
PhD research fellow @ McLean Hospital 🇺🇸 & University of Bergen 🇳🇴 | epigenetics in psychiatry 🧠🧬 | she/her
Postdoc at Neurocentre Magendie - French Institute of Health (INSERM) | Triathlete and runner.
https://joeribordes.github.io
PhD Candidate | Epigenomics | G x E | Stress Mechanisms | Max Planck Insitute of Psychiatry
PhD Student at the Regeneration and Neurogenomics Lab @ EPFL
Mainly here for #ComputationalBiology (long-term experience in academia, core facilities, small/medium companies and pharma), #Music and #Photography.
Machine Learning and Computational Biology at Helmholtz Munich, UZH, ETH Zurich, and previously at Genentech
Postdoctoral fellow, @MagdalenaGötz lab at @HelmholtzMunich & @BMC @LMU. Interested in cortical neurogenesis, its termination and environmental impact on it. 🇬🇷🇩🇪 alumnus of @Binder_Lab & @mpi_psychiatry, Sea admirer 🌅
Computational Biologist studying pancreas using single cell genomics.
PhD candidate at the Max Planck Institute of Psychiatry @binderlabmpip.bsky.social . Researching epigenetic signatures of early-life adversity. Passionate about stats and open science. 🧬🧠🏳️🌈
PhD Student in ML for Drug Discovery @TUM and Helmholtz Munich, prev: MILA, Oxford University, Heidelberg University - he/him
Postdoc between Helmholtz Munich and ETH Zurich | Theis Lab & Treutlein Lab | Computational Biology, Machine Learning & Organoids
[bridged from https://mastodon.social/@le_and_er on the fediverse by https://fed.brid.gy/ ]
Assistant Professor & Group Leader at @ki.se. Interested in prenatal environmental exposures and health risk and resilience in neuropsychiatry.
Official account of the Scientific Staff Association of
BSSE. Reposts are not endorsements.
We aim to better understand psychiatric disorders.
The international job board for scientists, by scientists | https://jobRxiv.org
Are you hiring? ➡️ https://jobrxiv.org/post-a-job
Get all new jobs in your inbox ➡️ https://jobrxiv.org/job-alert-subscription/
MD, Computational PhD in N Rajewsky Lab @ BIMSB-MDC, either sniffing around with my dog or listening cells chitchatting in the tumor microenvironment with high-resolution spatial transcriptomics
British/German biologist, biotechnologist & science manager. Grant manager @helmholtzai.bsky.social @www.helmholtz-munich.de. Enjoy science, travelling, sport, fine food & ale!