A huge thanks to all the co-authors Alex, Caterina, Chiara, Diogo, Russ and to @zkutalik.bsky.social for supervising this work. Also many thanks to the #Fulbright program for funding my research stay at Stanford where this work started.
#DBC @unil.bsky.social @sib.swiss #Unisanté
01.04.2025 20:12 — 👍 2 🔁 0 💬 0 📌 0
10/10 To conclude, we show that EHRs present great opportunities to study longitudinal phenotypes such as drug response. Although we found low heritabilities for PGx traits, there are more genetic predictors to be identified by combining resources and conducting GWAS in more diverse ancestries.
01.04.2025 20:12 — 👍 1 🔁 0 💬 1 📌 0

9/10 #PRS of the underlying biomarker level can predict drug efficacy, and high baseline cholesterol PRS were associated with increased absolute, albeit lower relative LDL reduction following statin treatment. However, PRS explained less than 2% of the variance.
01.04.2025 20:12 — 👍 0 🔁 0 💬 1 📌 0

8/10 More generally, a GWAS on cholesterol progression—defined as the difference between two cholesterol meas. in statin-free individuals —identified loci associated with cholesterol levels when adjusting for baseline meas. A GWAS on progression without baseline adjustment did not detect any signal
01.04.2025 20:12 — 👍 1 🔁 0 💬 1 📌 0

7/10 For example, in the analysis of LDL-C response to statin, the genetic association with the rs7412 SNP (APOE gene) is inflated when adjusting for baseline levels, since this SNP is also strongly associated with LDL-C baseline levels.
01.04.2025 20:12 — 👍 0 🔁 0 💬 1 📌 0

6/10 To avoid spurious associations with genetic variants that are also associated with baseline biomarker genetics, we propose a theoretical framework to model drug response and longitudinal phenotypes in an unbiased manner which implies not adjusting biomarker differences for baseline levels.
01.04.2025 20:12 — 👍 0 🔁 0 💬 1 📌 0
5/10 The contribution of rare variants to drug efficacy was found to be modest compared to common variants. We identified GIMAP5, a gene associated with autoimmune diseases, to impact absolute HbA1c response to metformin, however, the association did not replicate in the All of Us data.
01.04.2025 20:12 — 👍 0 🔁 0 💬 1 📌 0
4/10 In a systematic comparison with the literature, we find that GWAS derived from EHRs have comparable statistical power to GWAS conducted on PGx consortia and clinical trial data.
01.04.2025 20:12 — 👍 0 🔁 0 💬 1 📌 0

3/10 A major challenge with EHR data is that medication start, baseline and post-treatment measures are not reported as such. We tested multiple strategies and found prescription regularity to be important as well as the time between prescription start and the first post-treatment measure.
01.04.2025 20:12 — 👍 0 🔁 0 💬 1 📌 0

2/10 We then conducted #pharmacogenetic GWAS and rare variant burden tests. Discovery GWAS were conducted in the @ukbiobank.bsky.social and replication analyses in the #AllOfUSResearch. We don’t discover any new pharmacogenetic signals, but also don’t miss any major signal reported previously.
01.04.2025 20:12 — 👍 0 🔁 0 💬 1 📌 0
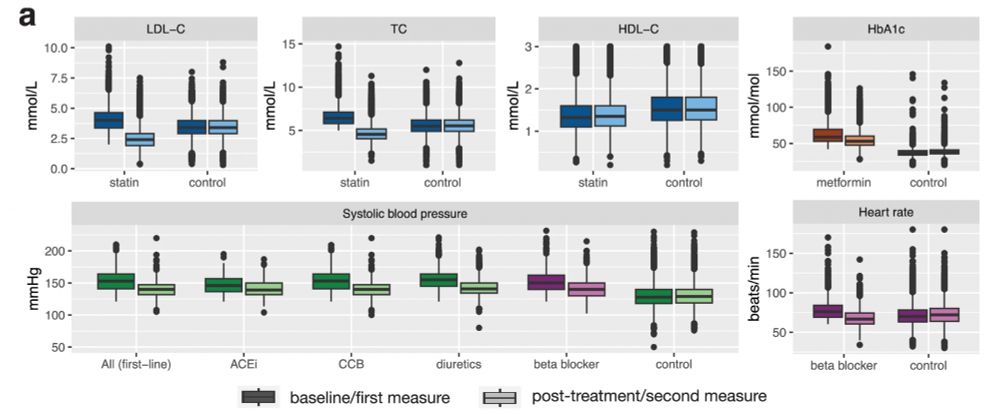
1/10 We identified drug regimens from medication prescriptions and extracted biomarker levels before and after drug initiation for ten cardiometabolic drug-biomarker pairs. For example, following statin treatment, LDL-C levels levels drop by ~40% which is below the levels of statin-free controls.
01.04.2025 20:12 — 👍 0 🔁 0 💬 1 📌 0

We study the genetics of drug efficacy using longitudinal biomarker and medication prescription data from #EHRs. 🧬💊
Have a look at the 🧵 on i) modelling drug response and biomarker progression, ii) using #PRS to predict efficacy and iii) the challenges with #EHRs.
www.nature.com/articles/s41...
01.04.2025 20:12 — 👍 11 🔁 4 💬 1 📌 1
Tissue-specific co-expression networks to identify key genes 🧬 Have a look at Patrick's thread that explains how Downstreamer can link common genetic variation to rare diseases. Thank you for including Zoltán and me on this project!
06.12.2024 13:25 — 👍 1 🔁 0 💬 0 📌 0
Computational medicine @ AU
Founder @ KH Biotechnology
Creator of spatiomic, pytximport and https://biocontext.ai
Interested in spatial omics, cancer & ageing
Lab: https://github.com/complextissue/
Personal: https://maltekuehl.com
Opinions my own.
CSEM is a Swiss technology innovation center developing advanced technologies with a high societal impact, which it then transfers to industry to strengthen the economy. The non-profit orientated, public-private organization is internationally recognized.
Protein Biochemist and Structural Biologist |
Postdoctoral Training Fellow at Sebastian Guettler’s group, the Institute of Cancer Research
| former: Low Group, Imperial College, Fasshauer Group, UniL and DCI Lausanne | #cryo-EM #teamtomo
MD-PhD bridging clinical medicine & AI/data science 🩺💻
Research Fellow @ Institute of Epidemiology and Prevention, Freiburg 🏥
🧬 Multi-omics data analysis and 🤖 Deep learning in genomics
"Attention Is All You Need" ⚡
https://orcid.org/0000-0001-8700-4335
Postdoctoral Fellow with Dr. Greg Gibson, Center for Integrative Genomics, Georgia Tech, Atlanta, GA
PhD Bioinformatics (Statistical Genetics) | MS Bioinformatics @GeorgiaTech
https://sininagpal.wixsite.com/snagpal
Welcome to the Institute for Stroke and Dementia Research (ISD) in Munich – Director: Prof. Martin Dichgans
https://www.isd-research.de/
• Now: postdoctoral fellow in gynaecological oncology at ISGlobal.
• Interests: environmental/social determinants; cancer; causal triangulation.
• Student: Public Health @lshtm.bsky.social; Theoretical Statistics @openuniversity.bsky.social.
The SIB Swiss Institute of Bioinformatics is an internationally recognized non-profit organization dedicated to biological and biomedical #DataScience.
We provide essential resources, expertise and services and represent Swiss bioinformatics.
Profil officiel de l'Université de Lausanne (Suisse)🇨🇭. "Le savoir vivant". Sept facultés. 17'000 étudiantes et étudiants. 15 programmes de bachelor, 42 programmes de master
Director/Founder, STEM Scholars Prog. Iowa State; STEM Equity speaker; Former Zoologist; Facilitator, SACNAS Leadership Institutes; Member of ◆Northern Cheyenne Nation◆ + 1st Gen/Pell; My views. I’m waay more fun live than online.
I like statistical genetics, forensic science, housing abundance (go YIMBY), getting around by bike & PT, sustaining democracy against plutocracy, fascism & Murdoch media, avoiding catastrophic climate change and drinking beer. Live in Melbourne, Australia
Nature Communications is an open access journal publishing high-quality research in all areas of the biological, physical, chemical, clinical, social, and Earth sciences.
www.nature.com/ncomms/
Bioinformatics Scientist / Next Generation Sequencing, Single Cell and Spatial Biology, Next Generation Proteomics, Liquid Biopsy, SynBio, Compute Acceleration in biotech // http://albertvilella.substack.com
Group leader in Statistical Genomics @fimm-uh.bsky.social
Lab website: https://www.helsinki.fi/en/researchgroups/computational-and-stat
Statistical Geneticist, Associate Professor @PittPublicHealth
#academicmom #statgen #genetics #education #rstats #biostatistics #likeablebadass #mothersleadingscience
Statistical Geneticist, Group leader at University of Lausanne/Unisante, father, climber, runner
Research fellow at Institute of Genetic and Biomedical Research, CNR | Cagliari







