BMC Biology is currently welcoming submissions of original research to the “Genome organization and evolution” Collection, which I am supporting as a Guest Editor.
If interested check out the link: www.biomedcentral.com/collections/...
Deadline: 26 March
#GenomeEvolution
@bmc.springernature.com
04.02.2026 11:19 — 👍 2 🔁 2 💬 0 📌 0
Congrats! 🙌
07.01.2026 20:22 — 👍 2 🔁 0 💬 0 📌 0
Congrats! 🙌
22.12.2025 23:28 — 👍 1 🔁 0 💬 0 📌 0
Anouk Necsulea, @maelledaunesse.bsky.social and myself are organising a symposium at SMBE this year.
Send us all of your abstracts!!
17.12.2025 11:17 — 👍 5 🔁 3 💬 0 📌 0

The tunicate Ciona - Nature Methods
The ascidian tunicate Ciona, one of the closest relatives of the vertebrates, inhabits shallow temperate waters in the worldwide ocean. A unique combination of simple stereotyped embryogenesis, regula...
Very happy to see this piece on the Tunicate Ciona published. Grateful to @alexandrejan.bsky.social and @chiaracastelletti.bsky.social for the Illustrations, and to @natmethods.nature.com for the opportunity to showcase our ever emerging model organism www.nature.com/articles/s41...
08.12.2025 18:33 — 👍 45 🔁 13 💬 0 📌 3
Thé Mercury effect
26.11.2025 11:46 — 👍 0 🔁 0 💬 0 📌 0
Cool!
18.11.2025 22:54 — 👍 1 🔁 0 💬 0 📌 0

Gene annotations matter for downstream analyses: different structural gene-annotation pipelines yield markedly different orthology calls. We compare four strategies across species and argue for standards & QC before inference. Work led by @silviaprietob.bsky.social. Paper: doi.org/10.1093/bioi...
23.10.2025 06:20 — 👍 13 🔁 5 💬 1 📌 0

Pleased to share our latest paper led by @tomlewin.bsky.social, now out in @currentbiology.bsky.social! We present the first chromosome-level genome of a phoronid and show that shared chromosomal fusions unite phoronids and bryozoans as sister groups.
www.cell.com/current-biol...
07.11.2025 16:20 — 👍 57 🔁 26 💬 7 📌 1

LondonEvoDevo
meeting's website
Come join the #LondonEvoDevo network half day meeting, hosted at @ucl.ac.uk on Friday November 7th, 2025. Submit your abstract by Oct 27th (or your interest in joining) here: forms.gle/TRbdrCkQTcY2.... Friendly vibes and free registration. More info here: londonevodevo.co.uk.
20.10.2025 13:42 — 👍 18 🔁 16 💬 0 📌 0
Congrats, even ALGs in it 😝!
17.10.2025 00:23 — 👍 1 🔁 0 💬 1 📌 0
bucolique!
02.10.2025 16:42 — 👍 1 🔁 0 💬 1 📌 0
Congrats!
30.08.2025 08:45 — 👍 1 🔁 0 💬 0 📌 0
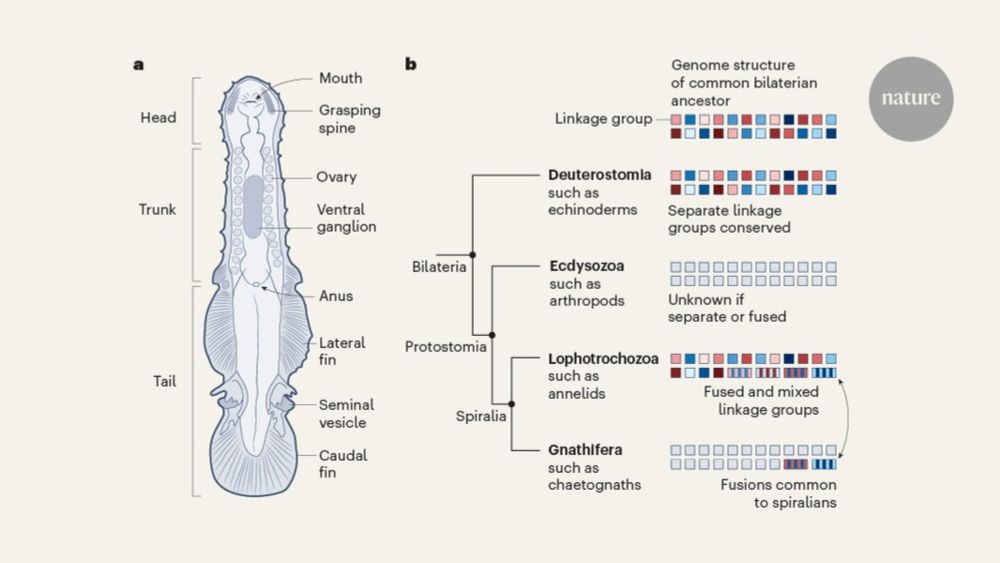
Chaetognaths have lost gene body methylation and shifted #5mC back to Transposable Elements. This reversion to the ancestral state is coupled with a simplification of DNMT3 architecture. We posit that trans-splicing might compensate.
It is an honour to have contributed to this 20y struggle ⛰️.
14.08.2025 07:53 — 👍 51 🔁 12 💬 2 📌 0

Cracking the Genetic Code of Arrow Worms: How Chaetognaths Got Their Unique Body Plan
New Nature study (co-authored by Queen Mary researchers) reveals that arrow worms evolved unique organs by inventing brand-new genes — not just reusing old ones. 🧬🌊
🔗 www.qmul.ac.uk/sbbs/news/it...
@qmulse.bsky.social
13.08.2025 15:16 — 👍 3 🔁 2 💬 0 📌 1
Finally, the enigmatic MedPost gene finds its place right where it belongs (between Meds and Posts) in an extended Hox cluster of >15 genes /5
13.08.2025 16:37 — 👍 6 🔁 0 💬 1 📌 0
We finally identified reversion to ancestral methylation patterns targeting mobile elements whose role in stabilising highly expressed genes was likely taken over by trans-splicing (hereby suggesting a possible role for this puzzling process) /4
13.08.2025 16:37 — 👍 5 🔁 0 💬 1 📌 0
We also confirmed the presence of spiralian ALG-fusions in chaetognaths despite fairly rearranged chromosomal architecture, as well as centromeres functioning without the classical CenH3 gene /3
13.08.2025 16:37 — 👍 5 🔁 0 💬 1 📌 0
We found that chaetognath genome is as puzzling as their body plan with extensive shuffling of gene repertoire, lots of gene novelty and loss, without evidence of WGD, which appear to be involved in the making of lineage-specific cell types /2
13.08.2025 16:37 — 👍 7 🔁 0 💬 1 📌 0
This surprisingly relaxing footage is from SIX MILES under the ocean – and it’s the deepest ecosystem yet discovered
31.07.2025 15:38 — 👍 14201 🔁 3422 💬 431 📌 526

Group Leader - Genome Biology Unit
Are you ready to lead groundbreaking research in Genome Biology? Join us at EMBL! We are seeking a motivated scientist to lead an independent research group addressing exciting and original biological...
To all post-docs: The Genome Biology dept @embl.org
has an Independent faculty position. Fantastic place to set up your lab –great package: core funding, fantastic Ph.D. students, cutting edge core facilities & great colleagues. Closing date Sept 19th
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
30.07.2025 13:41 — 👍 193 🔁 226 💬 0 📌 9

Repressive cytosine methylation is a marker of viral gene transfer across divergent eukaryotes
Abstract. Cytosine DNA methylation patterns vary widely across eukaryotes, with its ancestral roles being understood to have included both transposable ele
Happy to see our latest work out in @molbioevol.bsky.social. We revisit the evolution of 5-methylcytosine across neglected eukaryotic supergroups, establishing an ancestral repressive role silencing genome invaders, both transposons and viral elements👾: academic.oup.com/mbe/advance-... 🧵 1/7
28.07.2025 10:03 — 👍 101 🔁 51 💬 6 📌 0
Congrats!
24.07.2025 19:01 — 👍 1 🔁 0 💬 1 📌 0
Postdoctoral Researcher investigating sex chromosome evolution.
Bachtrog group | University of California, Berkeley
I am interested in Comparative and Evolutionary Genomics.
BS-MS Biology (IISER Berhampur)
Bioinformatics, Comparative Genomics, Phylogenomics, Evolutionary Genomics, Transcriptomics
Computational Biologist | Integrated PhD @IISER Berhampur | First-generation college graduate
Website: https://rohannathhere.github.io/Portfolio/
Postdoc@Okayama University, Ushimado Marine Institute / Evo-Devo / Morphology / Teleost fin morphology & gene regulation
PhD student (D3) | Evo-devo | fish fin development & evolution
https://researchmap.jp/kazuhide_miyamoto
Professor @Universitat Autonoma Barcelona-Understanding how genomes organize, evolve & are transmitted to the offspring. #genomics #evolution #meiosis #3Dgenome #biodiversity
Co-EiC @heredityjournal.bsky.social
👉https://webs.uab.cat/evolgenom/
Associate professor at the University of Barcelona.
Biologist
Evolutionary Biologist at UCSC | Assistant professor | Interested in genes, genomes, and #evolution of complex traits 🧬 | EvoDevo Regeneration
https://macias.sites.ucsc.edu/
📸 IG: @cnidolab_ucsc
Neurobiologist, Group Leader @msarscentre.bsky.social
https://www.chatzigeorgioulab.com/
Paramecium genome(s) aficionado; small RNAs, transposable elements, chromatin biology
PI at Institut Jacques Monod, Paris, France
Evolutionary cell biology, chromosomes, yeasts, and occasional SciArt🧬🧪🎨. Postdoctoral fellow at the labs of Gautam Dey (EMBL) and Gavin Sherlock (Stanford University).
EMBO postdoctoral fellow working in Marty Cohn’s lab at the University of Florida. From mouse genetics to evo-devo.
Postdoc at the University of Calgary | Ecology and Evolution | Bioinformatics
https://gabrielenocchi.weebly.com/
Research leader @NHM | Animal evolution | phylogenomics | molecular evolution
PhD-ing on Zebrafish development @fabianlab || Capturing life through lens and doodling are my favorite escapes..!!
Evolutionary genomics, nearly-neutralist...
Doctoral candidate in the Uri Frank Lab. Studying epigenetics in Hydractinia.
We are a lab focused on the study of invasive soil invertebrates, mainly land planarians. Led by Marta Álvarez Presas and based at the Universitat de Barcelona
lab at @UiTNorgesarktis studying microRNAs, paleotranscriptomics, biosystematics with a fable for flatworms.
MirGeneDB, MirMachine, MirMiner (soon!)