
ProFam: Open-Source Protein Family Language Modelling for Fitness Prediction and Design
Protein language models have become essential tools for engineering novel functional proteins. The emerging paradigm of family-based language models makes use of homologous sequences to steer protein ...
To advance the family-based modelling approach, we are releasing the entire framework open source:
ProFam Atlas: A curated, large-scale training corpus containing nearly 40 million protein families.
Code & Weights: github.com/alex-hh/prof...
Data: zenodo.org/records/1771...
22.12.2025 14:32 —
👍 3
🔁 1
💬 0
📌 0

For design, ProFam-1 excels at homology-guided generation. It produces diverse sequences with low sequence identity to natural proteins while preserving predicted structural similarity and conservation patterns of the natural family, even when conditioning on just a single example sequence.
22.12.2025 14:32 —
👍 2
🔁 1
💬 1
📌 0

By conditioning on homologous sequences, ProFam-1 is competitive with state-of-the-art zero-shot fitness prediction on ProteinGym, outcompeting much larger PLMs such as ESM.
22.12.2025 14:32 —
👍 2
🔁 0
💬 1
📌 0
Built by CATH, TÜM and NVIDIA, ProFam-1 is our new open-source protein family language model (pfLM) designed to generate functional protein variants and predict fitness using in-context example sequences.
22.12.2025 14:32 —
👍 11
🔁 5
💬 1
📌 1
Rob Finn on MGnify, everything bacteria and functions in different environments
16.09.2025 13:55 —
👍 2
🔁 1
💬 0
📌 0

Now Maria Martín from UniProt is telling us how AI-based tools are shaping the future of one of the key resources for protein sequences and function.
16.09.2025 13:34 —
👍 2
🔁 1
💬 1
📌 0

From structures to sequences, now Alex Bateman and the quest to annotate and classify all proteins!
16.09.2025 13:07 —
👍 2
🔁 1
💬 1
📌 0

Starting our afternoon session with a talk by Sameer Velankar, of PDBe and AFDB fame among other endeavours!
16.09.2025 12:46 —
👍 4
🔁 1
💬 1
📌 0

And now @gonzaparra.bsky.social on his first talk on protein frustration as a PI! Well done!
16.09.2025 11:32 —
👍 6
🔁 2
💬 1
📌 0

David Jones, on novel folds in AFDB and CATH’s founding being celebrated at a now-closed Pizza place in Euston Station
16.09.2025 11:09 —
👍 4
🔁 1
💬 1
📌 0

From CATH to Computational Enzymology, Dame Janet Thornton on the birth of CATH and beyond!
16.09.2025 10:44 —
👍 4
🔁 1
💬 1
📌 0

First Keynote by Burkhard Rost, on the impact of protein language models on the field of structural biology
16.09.2025 10:19 —
👍 4
🔁 1
💬 1
📌 0

Kickstarting our symposium “Protein Annotations in the age of AI” at UCL!
16.09.2025 10:19 —
👍 7
🔁 2
💬 1
📌 0
Congratulations @judewells.bsky.social!
22.08.2025 17:32 —
👍 5
🔁 2
💬 0
📌 0
If you'd like to showcase your research with a poster, details are included in the registration page.
We hope to see you there!
22.08.2025 10:45 —
👍 0
🔁 1
💬 0
📌 0
We have a stellar lineup of speakers!
Christine Orengo
Burkhard Rost
Janet Thornton
David Jones
Gonzalo Parra @gonzaparra.bsky.social
Sameer Velankar
Alex Bateman
Maria Martin
Rob Finn
Gerardo Tauriello
Alexey Murzin
22.08.2025 10:45 —
👍 3
🔁 2
💬 1
📌 0
There will be talks from world leaders in structural bioinfomatics on various themes including pioneering protein language models and key international resources including: PDBe, InterPro, UniProt, MGnify, SWISS-MODEL, FrustraEvo and CATH.
22.08.2025 10:45 —
👍 1
🔁 2
💬 1
📌 0

Protein Annotations in the age of AI
A not-for-profit symposium hosted at UCL - more details about speakers and venue below.
CATH turns 30 years old this year!
We are organising a 1-day symposium on September 16th at UCL, highlighting recent AI-based developments to enhance protein family classifications, annotations and analyses.
www.eventbrite.co.uk/e/protein-an...
22.08.2025 10:45 —
👍 12
🔁 7
💬 2
📌 0

Another CATH outing at Greenwich Park after a lovely cruise along the Thames and a pub lunch!
20.08.2025 18:18 —
👍 1
🔁 0
💬 0
📌 0

Metagenomic-scale analysis of the predicted protein structure universe
Protein structure prediction breakthroughs, notably AlphaFold2 and ESMfold, have led to an unprecedented influx of computationally derived structures. The AlphaFold Protein Structure Database now prov...
Our latest preprint is out on bioRxiv!
A collaboration between the groups of @martinsteinegger.bsky.social , David Jones and Christine Orengo, we clustered AlphaFold Database and ESMatlas, a whopping 821 million proteins!
We reveal biome-specific groups & over 11k novel domain combinations.
28.04.2025 11:16 —
👍 39
🔁 13
💬 2
📌 0
TED is a collaborative project between the structural bioinformatics groups of Professor David Jones & Professor Christine Orengo @cathgene3d.bsky.social at @ucl.ac.uk.
The TED integration is set to enhance the interpretability and usability of #AlphaFold predictions. Is this useful in your work?
03.03.2025 16:35 —
👍 10
🔁 2
💬 0
📌 0
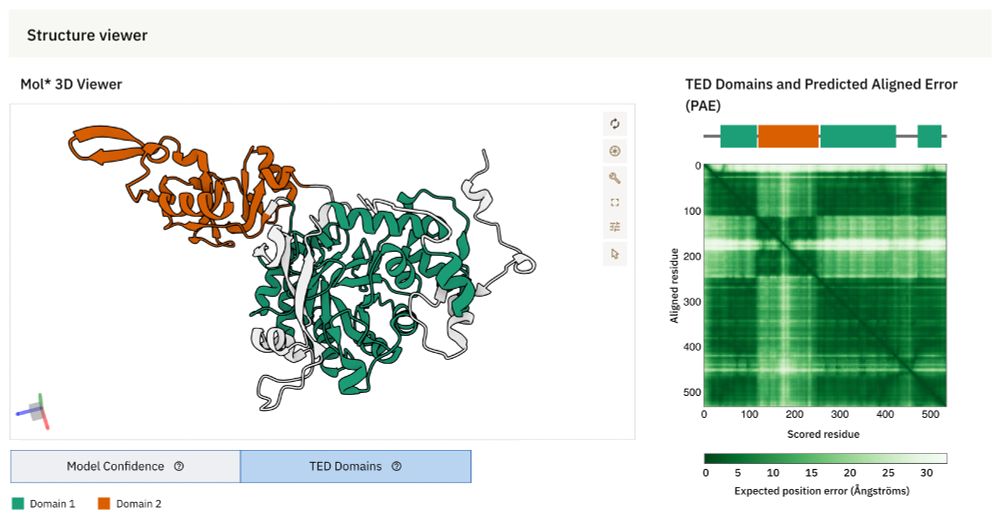
🚀 #AlphaFold Database update
AlphaFold DB now integrates The Encyclopedia of Domains (TED) – a resource designed to systematically identify & classify structural domains within AlphaFold-predicted protein structures.
www.ebi.ac.uk/about/news/u...
@pdbeurope.bsky.social
03.03.2025 16:33 —
👍 118
🔁 44
💬 1
📌 2


CATHmas lunch 2024!
18.12.2024 14:20 —
👍 1
🔁 1
💬 0
📌 0
We have updated sequences in our Functional Families by scanning FunFam-HMMs against UniProt release 2024_02, giving a 276% increase in FunFams coverage. The mapping of TED structural domains has resulted in a 4-fold increase in FunFams with structural information.
20.11.2024 12:52 —
👍 0
🔁 0
💬 0
📌 0
New PDB and TED data increases the number of superfamilies from 5841 to 6573, folds from 1349 to 2078 and architectures from 41 to 77.
20.11.2024 12:52 —
👍 0
🔁 0
💬 1
📌 0
CATH v4.4 represents an expansion of ∼64 844 experimentally determined domain structures from PDB. We also present a mapping of ∼90 million predicted domains from TED to CATH superfamilies.
20.11.2024 12:52 —
👍 0
🔁 0
💬 1
📌 0
We report a significant expansion of structural information (180-fold) for CATH superfamilies through classification of PDB domains and predicted domain structures from the Encyclopedia of Domains (TED) resource.
20.11.2024 12:52 —
👍 0
🔁 0
💬 1
📌 0

For those without access to the Science article, we added a full access link on the TED website (ted.cathdb.info) landing page!
6/6
16.11.2024 15:05 —
👍 0
🔁 0
💬 0
📌 0






















