And the legacy continues! 😊
@amjjbonvin.bsky.social @bioinfo.se @lindorfflarsen.bsky.social #EMBOIntegMod25 ! 🍀🧿
Büşra Savaş
@busrasavas.bsky.social
PhD student IBG & DEU | Computational Structural Biologist
@busrasavas.bsky.social
PhD student IBG & DEU | Computational Structural Biologist
And the legacy continues! 😊
@amjjbonvin.bsky.social @bioinfo.se @lindorfflarsen.bsky.social #EMBOIntegMod25 ! 🍀🧿
🚀 Excited to share that our article with @ezgikaraca.bsky.social is now published in Communications Biology!
In this study, we explored DNA readout rules of almost identical DNMT3A and DNMT3B (91% sequence similarity!), and we asked: how can nearly the same proteins “see” DNA so differently? 🧬✨
We just bumped into something very preliminary… but very exciting:
AF2.3 and AF3.0 distograms may potentially reproduce MD-like behavior.
Until we do further tests, you can reach our early insights at www.biorxiv.org/content/10.1...

Taken together, our results point to new exciting directions for structural biology through distograms:
- Rapid flexibility detection
- Improved EM map interpretation
- Distogram-informed ensemble modeling
Full story 👉 doi.org/10.1101/2025...
💻 GitHub: github.com/CSB-KaracaLa...

We next asked: how do AF2-based sampling strategies affect distograms?
We tested MinnieFold, AFsample2, and AF_cluster.
The biggest impact came from the AF2 version itself. Only AF2.3 mirrored AF3’s flexibility-aware distogram profile.
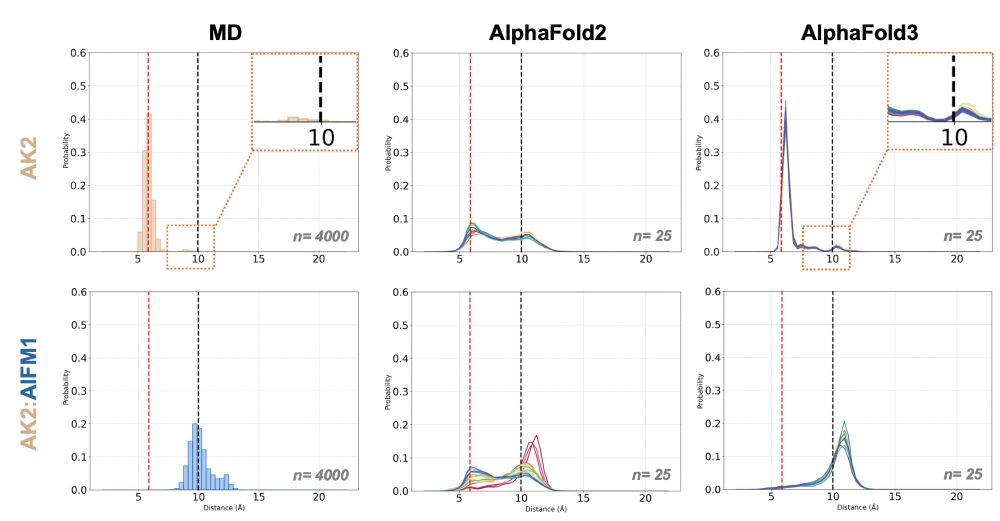
We run MD of apo and holo AK2 and revealed that holo AK2 hinge distance is sampled rarely in its apo state!
Strikingly, apo AF3 distogram reproduces the exact same behavior!!🤯
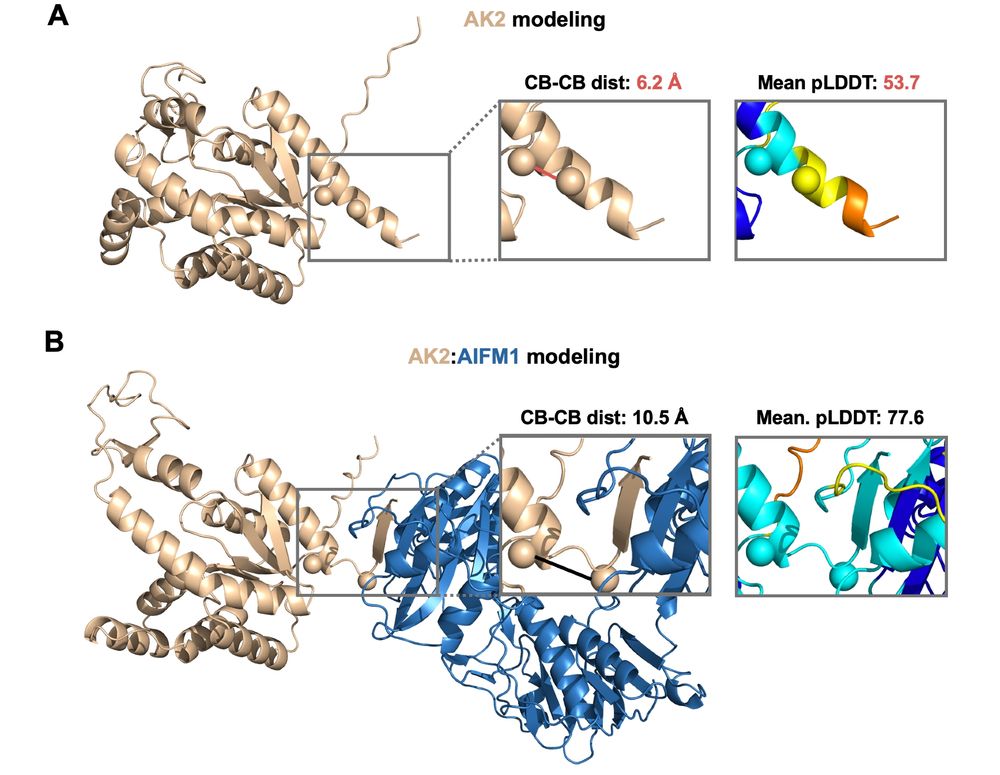
Surprisingly, AF3 apo and holo AK2 predictions beautifully reflect this conformational change, where C-ter of AK2 experiences a disorder-to-order transition upon beta-stacking itself to AIFM1👇
To track hinge motion, we concentrated on 230-233 CB–CB distances of AK2.
We tested this hypothesis on AK2:AIFM1 complex, a molecular switch between energy metabolism and cell death. 🔁💀
Its function depends on a hinge-driven motion, that is:
✔Recently revealed by two cryo-EM maps
✔ Occurs upon binding
❌Not seen in crystal structures

A primer on distograms: for each residue pair, AF outputs distance probability distributions.
Earlier, it was shown that the shape of the distogram peaks may inform us on protein flexibility.
How?
✔ Unimodal peak → rigid behavior
✔ Bimodal peak → flexible behavior
🚨 Super excited for our new preprint on flexibility cues in AlphaFold!
Together with @ezgikaraca.bsky.social and @aysebercinb.bsky.social, we found that distograms of AF2.3 and AF3 mirror MD sampling by predicting the extent of a novel conformational change! 🤯
For more👇

A small reminder to all structural biologists around working on biomolecular complexes: please consider sharing your complexes as targets for CAPRI - AI has not solved all structure prediction problems and there are still challenges! See www.capri-docking.org/contribute/
29.07.2025 06:26 — 👍 27 🔁 16 💬 2 📌 1
Only a few days left to apply for our EMBO course with a great lineup of speakers and tutors! Please feel free to RT!
Course website: meetings.embo.org/event/25-bio...
Organizers: myself, @amjjbonvin.bsky.social @bioinfo.se @lindorfflarsen.bsky.social
Location: www.ibg.edu.tr
It really was a blast!
22.05.2025 20:25 — 👍 1 🔁 0 💬 0 📌 0We want awe. We want discovery. And we want them now!
Huge thanks to Itai and Martin for the most enjoyable lecture on Night Science — a reminder that opennes lie at the heart of discovery!
This week's #SustainableTipTuesday is about computation - did you know there is a version of AlphaFold that uses 95% less energy than the regular version? Take a look below💡 #greenlabs
06.05.2025 09:00 — 👍 9 🔁 3 💬 1 📌 0
Inspired by Wallner's AFsample approach, @busrasavas.bsky.social's MinnieFold produced good quality antibody-antigen models even with 95% reduced sampling over CAPRI55 targets.
Just accepted in Proteins!
Paper: www.biorxiv.org/content/10.1...
Github: github.com/CSB-KaracaLa...

The legendary EMBO Integrative Modeling Course is back! If you want to be in this picture in the 2025 edition, just go ahead and register at meetings.embo.org/event/25-bio...
Organized by myself & @amjjbonvin.bsky.social & @arneelof.bsky.social & @lindorfflarsen.bsky.social
Super proud to have been listed as one of the successful groups in predicting the structure of an antibody-peptide during @capridock.bsky.social's 55th round! This was our first attempt in CAPRI as the Karaca team with @busrasavas.bsky.social @iremylmazbilek.bsky.social @atakanozsan 💪
13.01.2024 09:21 — 👍 11 🔁 2 💬 1 📌 0