

@rowancallumg.bsky.social and @liyamiksovsky.bsky.social demonstrating our newest lab tradition - creating virus snowflakes (designed by @socialinfluenza.bsky.social ) out of submitted grants and fellowships.
🧪 #socialviruses
@rowancallumg.bsky.social
Postdoc studying viral evolution with @asherleeks.bsky.social | previously @mermanchester.bsky.social | mutagenesis | bacteria | viruses | big R nerd 🧬🦠💻 🏳️⚧️ He/Him


@rowancallumg.bsky.social and @liyamiksovsky.bsky.social demonstrating our newest lab tradition - creating virus snowflakes (designed by @socialinfluenza.bsky.social ) out of submitted grants and fellowships.
🧪 #socialviruses

@rowancallumg.bsky.social @knightjar.bsky.social @rokkrasovec.bsky.social et al. study the antimutator ΔnudJ to highlight the value of considering mutation rate and mutational spectrum to understand the evolution of mutation rate-modifying alleles.
🔗 doi.org/10.1093/molbev/msaf182
#evobio #molbio

Across domains of life 2017: doi.org/cb9s
Potential mechanism 2024: doi.org/m8zt
As discussed in the 2024 paper, ecological relevance likely depends on the amount of through-flow in the environment. Less flow = more opportunity for collective detoxification of the environment
Great thread! All good, but this does grab the attention:
"our findings with ΔnudJ suggest future anti-evolution drug strategies could suppress spontaneous resistance evolution not only through minimizing resistance mutations but also by specifically limiting access to the fittest mutations."

Finally, a shoutout to the blueycolors colour scheme for making all of my ggplots far more fun! 🟦 🟧 13/13
18.08.2025 15:56 — 👍 2 🔁 0 💬 0 📌 0Deleting the nudJ gene not only makes rifampicin resistance less likely by reducing the mutation rate but also reduces the chance of a resistant mutant rising to fixation as ΔnudJ favours lower fitness mutations: synergystic effects of mutation rates and spectra impeding resistance evolution 12/13
18.08.2025 15:54 — 👍 0 🔁 0 💬 1 📌 0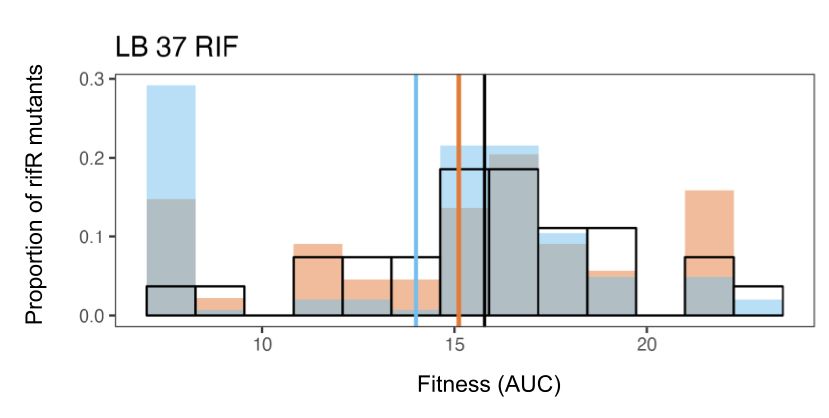
Distribution of fitness effects showing mean fitness to be lowest in nudJ knockout followed by the wt followed by the null expectation of all mutations as equal rates
Resistance mutations accessed by ΔnudJ (blue) were significantly less fit than those accessed by the wildtype (orange) & both strains accessed mutants less fit than one would expect by chance (black: all mutations occurring at =rates): spontaneous mutation is not optimised for future fitness. 11/13
18.08.2025 15:53 — 👍 0 🔁 0 💬 2 📌 0Rather than looking at a genome-wide scale we took rifampicin resistance mutants observed in each genetic background and measured their fitness in a common genetic background. This choice to consider fitness at a single locus is reflective of resistance evolution to many antibiotics. 10/13
18.08.2025 15:51 — 👍 0 🔁 0 💬 1 📌 0Now for the cool evolution part: work from @deepaagashe.bsky.social and others has shown that shifts in the mutational spectrum can lead to genome wide changes in the fitness effects of accessed mutations. Was our ΔnudJ strain accessing mutations with different fitness effects to the wildtype? 9/13
18.08.2025 15:50 — 👍 0 🔁 0 💬 1 📌 0
Proportion of rifR mutations accounted for by each SNP type (+small indels) showing nudJ knockout to have less A>G and more A>C mutations, proportionally, than the wt
A mutation rates is made up from the rates of many different types of mutations (the mutational spectrum). We find that ΔnudJ has a significantly altered mutational spectrum to the wildtype, e.g. in the wt 22% of mutations are A>G, in ΔnudJ this is 9%. This is reversed by IS1 mutation in waaZ. 8/13
18.08.2025 15:49 — 👍 0 🔁 0 💬 1 📌 0
Scatter plot of mutation rate as a function of pop density showing nudJ knockout to have lower mutation rate and less variation by density while wt and nudJ + secondary mutation strains have high mutations rates significantly negatively correlated with pop density.
The antimutator ΔnudJ not only reduces mutation rates but also decreases the responsiveness of mutation rates to population density (usually mutation rates are elevated at low population densities in batch culture). Both of these traits are reverted by our strains carrying secondary mutations. 7/13
18.08.2025 15:48 — 👍 0 🔁 0 💬 2 📌 0I then focussed in on ΔnudJ, finding surprisingly that the mutation rate had reverted to the of the wildtype! We found that independent mutations in 2 of our stocks were responsible for this phenotype reversion, with initially frustrating inconsistencies resulting in new genetic understanding. 6/13
18.08.2025 15:46 — 👍 1 🔁 0 💬 1 📌 0
Mutation rates of nudix knockouts, mutT is the only mutator (mut rate > wt) and nudHJFB are antimutators (mut rate < wt)
Disruption of nucleotide metabolism gene mutT increases mutation rates however, its sibling genes in the nudix hydrolase family had not been characterised. A massive survey by coauthor Huw Richards found that 4 knockouts from this family result in significant mutation rate decreases (nudBFJ&H) 5/13
18.08.2025 15:45 — 👍 0 🔁 0 💬 1 📌 0Mutator strains in which disruption of a single gene (usually DNA replication and repair enzymes) increases mutation rates are very common in laboratory and clinical E coli populations. BUT gene disruptions that reduce mutation rates (antimutator strains) are far less frequently observed (4/13)
18.08.2025 15:44 — 👍 0 🔁 0 💬 1 📌 0The antibiotic killed cells could stretch from Manchester to Paris and back to the UK while the total cell count would reach from my PhD lab in Manchester @mermanchester.bsky.social to my postdoc lab in Vancouver @zoology.ubc.ca 4.7 times. (3/13)
18.08.2025 15:43 — 👍 0 🔁 0 💬 1 📌 0This team effort involved 4 PhD students, 1 undergrad, 1 masters student, 6 groups leaders, 7 years of (on-and-off) research, >8.5x10^11 E. coli cells killed with antibiotics and >3.5x10^13 E. coli cells grown in total. (2/13)
18.08.2025 15:41 — 👍 0 🔁 0 💬 1 📌 0
New paper out @molbioevol.bsky.social on antimutator Ecoli, mutational spectra and the distribution of fitness effects of antibiotic resistance with @knightjar.bsky.social @rokkrasovec.bsky.social and others @mermanchester.bsky.social doi.org/pzfm
Read🧵for highlights and fun stats (1/13)
🦠 New paper led by James Horton (feat. J Cherry & @gretelwaugh.bsky.social) reveals GnT DNA motif can boost T→A mutation rates up to 1000×, & how tweaking nearby bases can fine-tune its potency.
General across bacteria + potential in synbio.
📄 academic.oup.com/mbe/advance-ar…
🔗 James’s thread ⬇️

Aerobic bacteria show higher A>G (and lower G>A) mutation rate than anaerobes. First results in bioinformatic project to estimate mutation spectra across bacterial tree under my supervision. Qs and ideas are wellcome. #SMBE2025
22.07.2025 13:12 — 👍 27 🔁 6 💬 2 📌 0Amazing conference
27.05.2025 02:26 — 👍 2 🔁 0 💬 0 📌 0Same genome, different shape, different outcome.
SOS-independent phenotypic differences shape survival in E. coli.
Very happy to share our preprint!


Empty desk in Manchester ready for new adventures in microbial evolution elsewhere 🤯🦠 I can't express how much I will miss everyone in @mermanchester.bsky.social (leaving was made a little sweeter by my first ever 'best talk' prize at #northernBUG / #YUEG at @huddersfielduni.bsky.social last week)
31.03.2025 18:26 — 👍 15 🔁 0 💬 5 📌 1
We have a 2-year postdoc position available in the Lind Lab at Umeå University, Sweden! Predicting and steering experimental evolution of antibiotic resistance in Pseudomonas aeruginosa. Reposts appreciated.
umu.varbi.com/en/?jobtoken...
Thank you so much to my examiners for a great discussion. And to my supervisors and lab mates for making the past 4 years such a positive and formative experience! @mermanchester.bsky.social is the place to be ❤️ (also thank you to e coli for mutating every day, all of your mutations are appreciated)
19.02.2025 15:10 — 👍 13 🔁 0 💬 0 📌 0
Glad to see this out today in MEE!
The rarity of mutations and the inflation of bacterial effective population sizes
besjournals.onlinelibrary.wiley.com/doi/10.1111/...
#popgen #populationgenetics #evolution #bacteriagenomics #bacteria #microsky #statsstandrews @uniofstandrews.bsky.social

Excited to speak at #PGG58 / #PopGroup58 about mutation rates, mutation spectra, antibiotic resistance and (if I talk fast enough) how the environment affects these.
Tuesday in LT3 right after lunch. 🦠🧬
Come for the mutations, stay for the Bluey colour schemed ggplots. 📊🟦🟧
Fully funded PhD project on single-molecule dynamics of phages & plasmids.
Join us and learn a broad range of skills, from synthetic biology and evolution to 3D microscopy.
#AMR #microscopy
Please contact me directly if you wish to discuss informally and please repost!
tinyurl.com/mrjmze7j
Amazing project and supervisors!!! 🔬🦠🧑🔬
18.12.2024 14:15 — 👍 2 🔁 2 💬 1 📌 0
🚨 The Whelan lab is hiring! 🚨 We're looking for a Research Assistant to join our lab at U of Manchester. We're seeking someone with experience in microbiology & microbiome to support our growing group. Note this isn't a postdoc role (though keep 👀 for those announcements soon!) #Microsky 🧪 🧬 💻
24.09.2024 08:04 — 👍 33 🔁 44 💬 0 📌 2
1st foray into T6SS with talented @mermanchester.bsky.social fellow Will Smith: eco-evo models and experimental evolution show how and why producing multiple toxins blocks resistance evolution (even though cross-resistance can evolve under single-toxin attack) www.biorxiv.org/content/10.1...
31.07.2024 17:17 — 👍 28 🔁 19 💬 1 📌 1