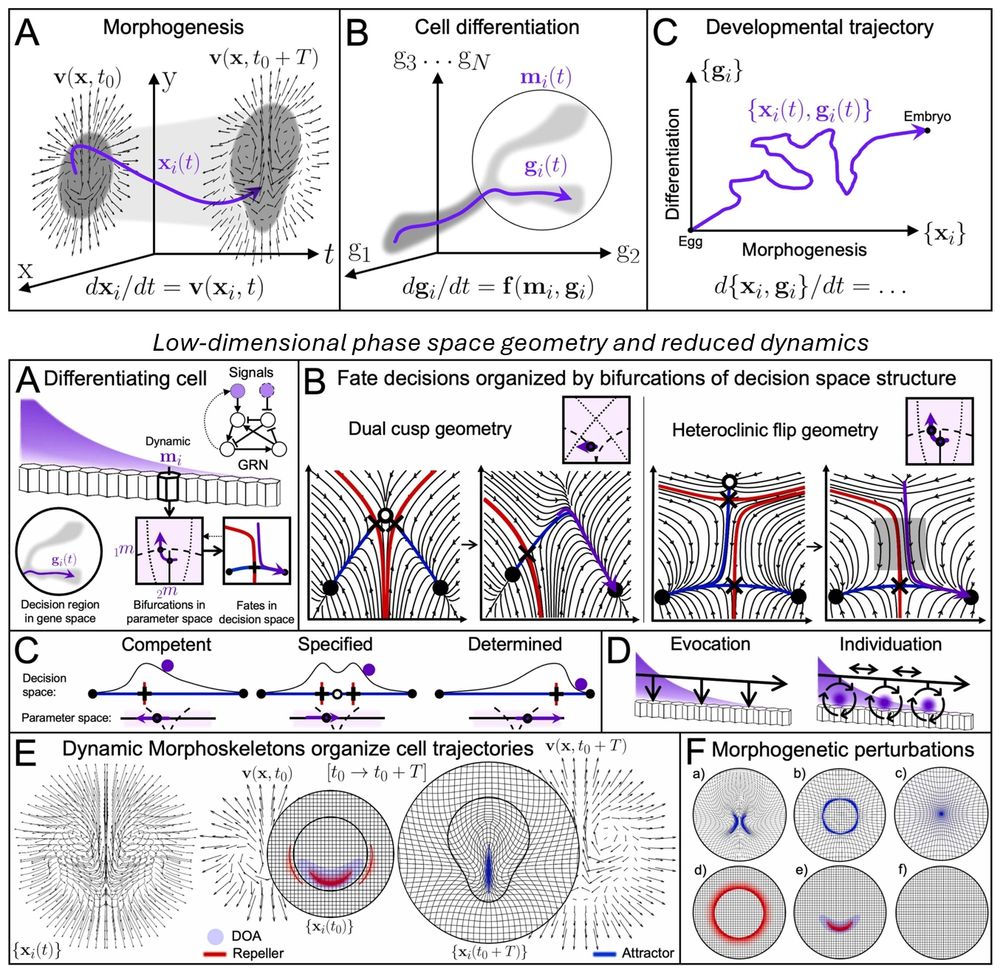
📣 New review! Dynamical systems and low-dimensional geometric structures in phase space help rationalize how embryos develop form and function, from large datasets. We focus on morphogenesis, cell differentiation, and their interconnection.
@alex-plum.bsky.social
sciencedirect.com/science/arti...
04.06.2025 16:46 — 👍 44 🔁 9 💬 0 📌 3

Are you passionate about the mathematics of data science, computational geometry and topology and want to apply your skills to a beautiful biological question?
Come do a PhD with us!
You will help us understand of how boundaries are established and maintained in early heart development.
15.11.2025 15:41 — 👍 5 🔁 6 💬 1 📌 0
... and the application process: centuri-livingsystems.org/centuri-phd-...
If you want to develop new quantitative methods, collaborate across disciplines, and work on an exciting biological problem, we’d love to hear from you!
15.11.2025 15:41 — 👍 0 🔁 0 💬 0 📌 0
You will join the collaborative, interdisciplinary environment of the Turing Centre for Living Systems (CENTURI) in Marseille, working within the teams of Paul Villoutreix and Robert Kelly!
The full project: centuri-livingsystems.org/wp-content/u...
15.11.2025 15:41 — 👍 0 🔁 0 💬 1 📌 0

Are you passionate about the mathematics of data science, computational geometry and topology and want to apply your skills to a beautiful biological question?
Come do a PhD with us!
You will help us understand of how boundaries are established and maintained in early heart development.
15.11.2025 15:41 — 👍 5 🔁 6 💬 1 📌 0
Felix Zhou's u-Segment3D is out:
www.nature.com/articles/s41...
It leverages 2D cell segmentations from orthoviews to generate a ‘consensus’ 3D segmentation.
It does a great job on segmenting densely packed samples, and we have used the algorithm in many applications.
12.11.2025 02:41 — 👍 78 🔁 18 💬 1 📌 0

Congratulations to Dr @baalberti.bsky.social !! 🎉
And many thanks to his jury members @randersson.bsky.social @arnausebe.bsky.social @olivier-gandrillon.mastodon.social.ap.brid.gy Marie Sémon Anouck Necsulea and co-supervizor @paulvilloutreix.bsky.social
10.11.2025 09:37 — 👍 12 🔁 2 💬 1 📌 0
Excited to be in Paris later this week!
09.11.2025 09:37 — 👍 2 🔁 0 💬 0 📌 0

#Agenda
With
@batistaruan.bsky.social, Valentin Rineau, @ignacioq.bsky.social, Aleksandra Walczak, Guillaume Achaz, @k4tj4.bsky.social - @r3rto.bsky.social, Maël Montévil, @paulvilloutreix.bsky.social, Barbara Bravi, Cyril Rauch, Giuseppe Longo, Anton Robert, @charbelelhani.bsky.social , Ana Soto
09.11.2025 08:36 — 👍 5 🔁 3 💬 0 📌 1
Great program, amazing scientists and beautiful facilities!! I visited for 6 months in 2023, a unique place in Europe at the intersection of maths, computer science and developmental biology
31.10.2025 13:46 — 👍 4 🔁 1 💬 0 📌 0

🧬 Excited to share Nicheformer out now in Nature Methods!
A transformer foundation model linking single-cell & spatial omics, learning spatial context from gene expression to map tissue organization.
Led by Ale Tejada & Anna Schaar 👏
👉 www.nature.com/articles/s41...
30.10.2025 19:19 — 👍 28 🔁 7 💬 0 📌 0


Had a very nice time visiting Granada for the annual heart ICDAR meeting! We presented Harshit Pateria's current work in collaboration with Robert Kelly following up from our recent publication on the second heart field www.nature.com/articles/s41...
17.10.2025 15:51 — 👍 4 🔁 1 💬 0 📌 0
Wow @gioelelamanno.bsky.social & @giodang.bsky.social
16.10.2025 07:09 — 👍 12 🔁 2 💬 0 📌 0


Pleasure to announce the program of my new series of lectures at the College de France in Nov-Dec. Continuing on the theme of Biological information. I will focus on the computational aspects, building on David Marr's tri-level of analysis in biological systems. YouTube link sent when it starts.
07.10.2025 06:12 — 👍 34 🔁 11 💬 1 📌 0
Very elegant and efficient methodology to recover spatial patterns from multi-modal spatial datasets 👌!! Many congratulations @paulvilloutreix.bsky.social and to all authors👏😀!!
19.09.2025 09:16 — 👍 3 🔁 1 💬 0 📌 0
jsPCA is simple, it is based on the product between the gene expression covariance (classical PCA) and the spatial autocorrelation.
We have shown that it's fast, interpretable and highly adaptable to multiple settings and large datasets.
Congrats to Ines Assali who led the project!
19.09.2025 06:49 — 👍 2 🔁 0 💬 1 📌 0
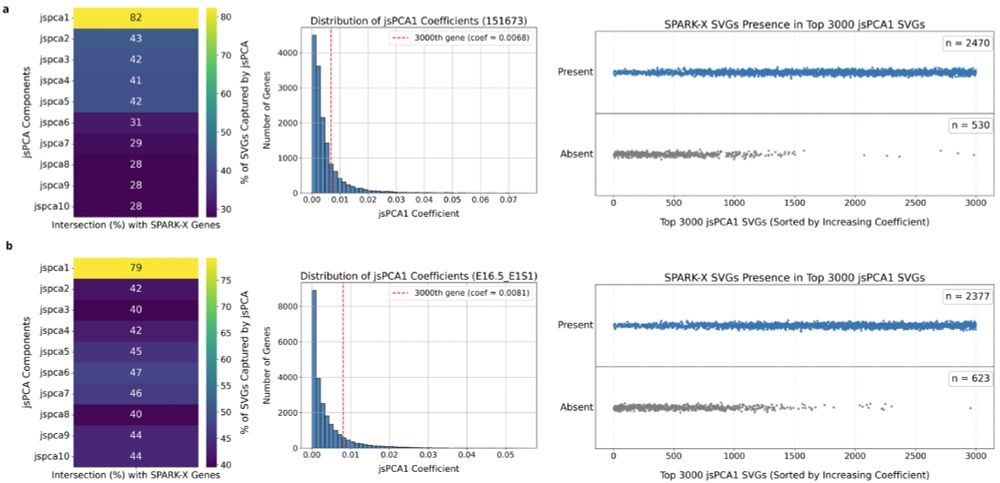
Finally, principal components of jsPCA are directly interpretable in terms of spatially variable genes (SVG). We found that the top 3000 genes of the first principal component recovered 80% of the SVGs obtained by the widely used SPARK-X method.
19.09.2025 06:49 — 👍 0 🔁 0 💬 1 📌 0

jsPCA can also learn a joint representations on some of the slices and use it to predict the domains in an unseen slice.
19.09.2025 06:49 — 👍 0 🔁 0 💬 1 📌 0


The joint analysis of multiple slices by jsPCA generates common domains among datasets, in contrast to monoslice analysis..
19.09.2025 06:49 — 👍 0 🔁 0 💬 1 📌 0


jsPCA achieves state-of-the-art accuracy for domain identification while reducing computation from hours to seconds, and scales to atlas-level datasets such as Stereo-seq.
19.09.2025 06:49 — 👍 0 🔁 0 💬 1 📌 0

Very happy to introduce jsPCA, a fast and interpretable computational framework for spatial transcriptomics that simultaneously identifies spatial domains and variable genes across multi-slice and multi-sample data.
19.09.2025 06:49 — 👍 17 🔁 8 💬 2 📌 0

hoxd13a reporter in a zebrafish larva
Are digits modified fins, or evolutionary innovations? Read how we tackled this old question from a new angle🧪
A story with @chasebolt.bsky.social, @homeobox.bsky.social and myself, coordinated by @denisduboule.bsky.social from @college-de-france.fr and published in @nature.com today!
#InHoxWeTrust
17.09.2025 17:26 — 👍 111 🔁 44 💬 4 📌 6
Cosmologist, pilot, author, connoisseur of cosmic catastrophes. TEDFellow, CIFAR Azrieli Global Scholar. Domain verified through my personal astrokatie.com website. She/her. Dr.
Personal account; not speaking for employer or anyone else.
CNRS Researcher at the Institut de Biologie de l’École Normale Supérieure, Paris.
Studying evolutionary processes using stochastic phylogenetic models.
Failed reincarnation of Montaigne.
Group leader CRG; Associate Faculty ToL Sanger Institute.
Genome regulation, chromatin, cell types, and evolution. https://www.sebepedroslab.org
Comparative developmental biology, regeneration, non-conventional model organisms, live imaging; see www.averof-lab.org
Group Leader (Amidex Chaire d'Excellence) at DyNaMo, Aix-Marseille Université @dynamo-lab.bsky.social @univ-amu.fr
Biophysics theory, statistical physics
Developmental biologist interested in embryonic self-organization.
Group leader at Institut Pasteur in Paris, France.
International Conference on Learning Representations https://iclr.cc/
Doing a Ph.D. AI in Bio. | Ex @WhiteLabGx @BroadInstitute @MIT | Built @PiPleteam | ML, Cancer, Genomics, Data Sci, Entrepreneur, FullStack Dev | All views are mine
Associate Professor of Statistical Genomics (Oxford Stats)
https://www.stats.ox.ac.uk/~ignatiev/
Assistant Professor at Ecole Polytechnique, IP_Paris// Before: Oxford_VGG, Inria Grenoble // multimodality, genAI enthusiast // happy mum+dog_mum // opinions: mine
Asst Professor at Johns Hopkins (AMS and DSAI). Previously: Simons Institute, Oxford stats, Polytechnique. I like to scale up things!
https://www.soufianehayou.com/
The Erlangen AI Hub brings together leading minds from across the UK’s mathematical, algorithmic and computational communities. We employ foundational tools to break new ground in AI, and redefine its future use to benefit science, industry and society.
Bringing together Gen AI researchers to collaborate on models driving impacts for science, industry & society. https://linktr.ee/genaihub
We're the Department of Statistics at the University of Oxford (UK). We provide teaching & complete research on computational statistics and statistical methodology, probability, bioinformatics and mathematical genetics.
https://www.stats.ox.ac.uk/
Official account of the Mathematical Institute, University of Oxford
Department of Computer Science at the University of Oxford, sharing news on our outstanding research across a broad spectrum of computer science #CompSciOxford
Biophysics, developmental biology, morphogenesis
@ IBDM & Centuri (CNRS / Aix-Marseille Université)
https://raphaelclement.wordpress.com/
Professor of Applied Physics at Stanford | Venture Partner a16z | Research in AI, Neuroscience, Physics
Bioinformatics, Comp biol, cancer genomes, text mining, strucutral bioinfo. but also: Science publishing, science policy, genome & ethics, science & art
PI at Institut Pasteur
Evolution, immunity, genomics, microbiolgy.
Into immunity in bacteria and its conservation in eukaryotes.
Advocate for more inclusive sciences
https://research.pasteur.fr/en/team/molecular-diversity-of-microbes/

























