
How embryos stay “on time” as they grow?
Happy to share our recent publication: Quantitative modeling of mRNA degradation reveals tempo-dependent mRNA clearance in early embryos
(1/7)
academic.oup.com/nar/article/...

How embryos stay “on time” as they grow?
Happy to share our recent publication: Quantitative modeling of mRNA degradation reveals tempo-dependent mRNA clearance in early embryos
(1/7)
academic.oup.com/nar/article/...
1. Today the NIH director issued a new directive slashing overhead rates to 15%.
I want to provide some context on what that means and why it matters.
grants.nih.gov/grants/guide...
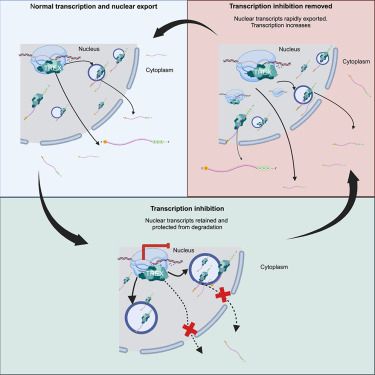
Why can a human tolerate a drug that globally inhibits transcription? Why do transcription inhibitors not cure cancer? Our first paper of 2025 may help explain (some) of this!
So incredibly proud of @tobiaswilliams.bsky.social & Ewa Michalak who led the work!
www.cell.com/molecular-ce...
Ever wondered how transcription choreographs histone modifications? Our work reveals the basis of co-transcriptional H3K36me3 by SETD2. We visualize how a histone writer coordinates with the transcription machinery! This is the magnus opus of @jonmarkert.bsky.social!
tinyurl.com/setd2

Have you been thinking hard about statistical modelling of scATAC-seq data? (No.)
Luckily for you, @aaronkwc.bsky.social has!
Aaron will help you grok:
What's going on?
What is TF-IDF?
Is there really single-cell level chromatin information?
Check it out 👇
www.biorxiv.org/content/10.1...
🧪🧬💻
www.biorxiv.org/content/10.1...
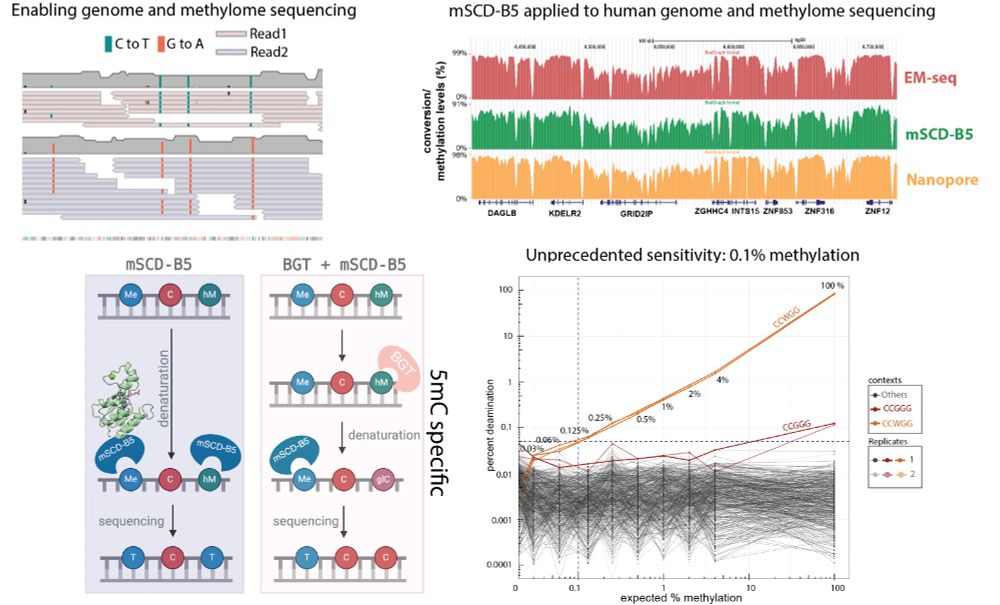
Our study published today on #bioRxiv describe the identification of a deaminase that converts 5mC to T, enabling direct sequencing of the human methylome and genome. This achievement was made possible through a collaborative effort across all departments at #NEB.
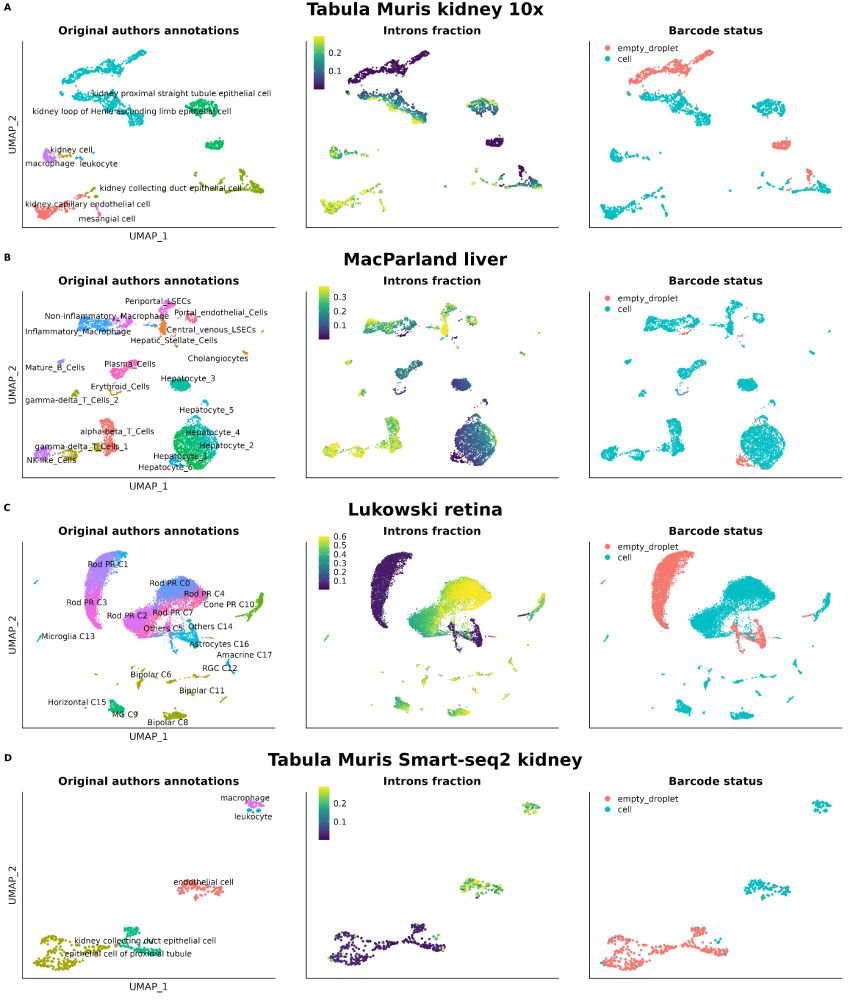
Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8
There's been some interest in this PLOS announcement, so despite it being light on details, here's why it's big. In our recent study 'The strain on scientific publishing' we showed how certain groups took off in total articles published.
Why? And why now?
direct.mit.edu/qss/article/...
1/n 🧵

🧬🔍There are 50 petabases of freely-available DNA sequencing data. We introducing Logan Search which allows you to search for any DNA sequence in minutes, bringing Earth’s largest genomic resource to your fingertips.
🏔️ logan-search.org 🏔️
#Genomics #Bioinformatics #OpenScience

Today in Molecular Cell the work of one of my favorite (ex)PhD students in the Wysocka Lab: Dr. Christina Jensen. I saw this project develop throughout the years and it has kept me engaged in hypothesizing scenarios in every lab meeting. Congratulations to the authors!
www.cell.com/molecular-ce...

Enhancer cooperativity can compensate for loss of activity over large genomic distances | Molecular Cell
#chromatin #enhancers
www.cell.com/molecular-ce...
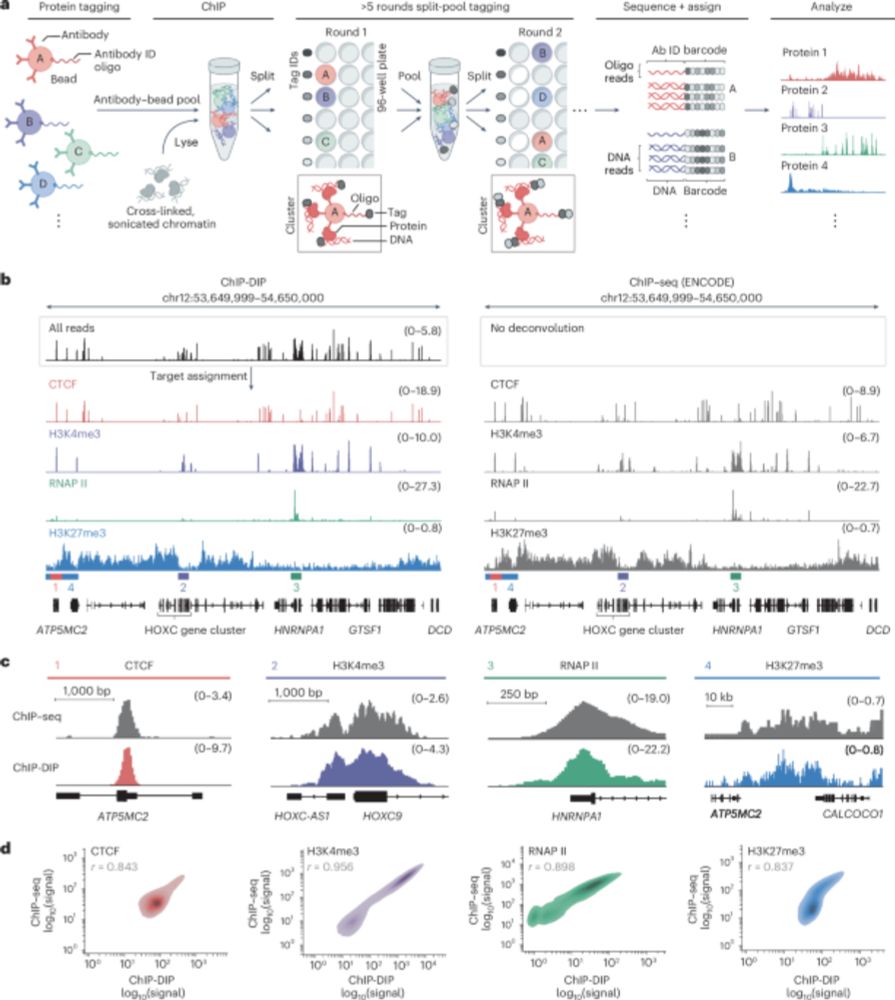
Gene regulation involves thousands of proteins that bind DNA, yet comprehensively mapping these is challenging. Our paper in Nature Genetics describes ChIP-DIP, a method for genome-wide mapping of hundreds of DNA-protein interactions in a single experiment.
www.nature.com/articles/s41...
Here’s the updated Computational Biology Starter Pack! Let me know if you'd like to be included.
go.bsky.app/QVPoZXp

Evolving cell states and oncogenic drivers during the progression of IDH-mutant gliomas - Nature Cancer 🧪
www.nature.com/articles/s43...
Hi, can you please add me? Thank you, Yotam.
24.11.2024 05:58 — 👍 0 🔁 0 💬 0 📌 0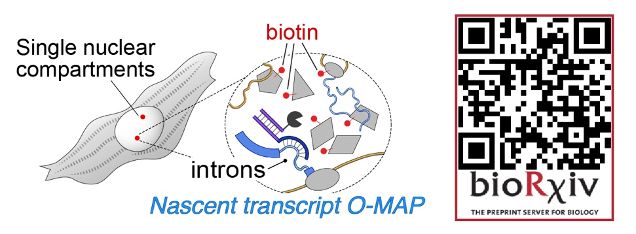
It's my great pleasure to present the next big preprint from SheqLab! An exciting application of our O-MAP platform that I hope will transform the study of nuclear architecture.
If you've ever wanted to dissect the subnuclear "neighborhood" around an individual locus, read on! (1/30)

Plotting becomes tidy in R , good read for trainees.
www.biorxiv.org/content/10.1...
Our paper with Carmel lab, the work of PhD student Yoav Mathov was just published in Nat Ecol Evol. www.nature.com/articles/s41....
Inferring DNA methylation in non-skeletal tissues of ancient specimens. A major challenge in studying DNA methylation in ancient samples is that it is tissue-specific.
Leonid Mirny and I wrote this for all interested in chromosomes: "The chromosome folding problem and how cells solve it"
www.cell.com/action/showP...
Great list! Can you add me too? Thanks.
17.11.2024 21:17 — 👍 1 🔁 0 💬 1 📌 0I’ve started making a starter pack for computational #cancer! Let me know if you’d like to be added or know someone who should be go.bsky.app/N6moBmG
12.11.2024 20:58 — 👍 43 🔁 18 💬 24 📌 0
Eden showed how SMCHD1 regulates the splicing of DNMT3B1, and how the promoted isoform changes DNA methylation at the D4Z4 satellite repeat to induce DUX4 expression, explaining how SMCHD1 mutations lead to FSHD. More details at: dx.doi.org/10.1126/scia...
#Epigenetics #Splicing #SMCHD1
Importing some of our recent updates from Twitter to facilitate the great migration. Happy to report our results on the role of SMCHD1 in facioscapulohumeral muscular dystrophy (FSHD). A very close and fun collaboration with Prof. Salton, this work was led by our amazing joint student, Eden Engal.
17.11.2024 15:40 — 👍 4 🔁 1 💬 1 📌 0
Importing some of our recent updates from Twitter to facilitate the great migration. Interested in inferring pathway activity in the single cell level from #SingleCell RNAseq data? check out SiPSiC. Details at: t.co/zeLafAiMHg
Available on bioconductor: t.co/CyK0YVokV9 and github: t.co/EzSWAUFeel

EpiSci -
www.cell.com/molecular-ce...

(1/8) 🚀 Excited to share our findings on a large-scale ChIP-seq assay for 166 previously uncharacterized human transcription factors (TFs) and their roles in both regulatory regions, and more strikingly, the “dark matter” genome. 🌌 doi.org/10.1101/2024....
15.11.2024 19:01 — 👍 93 🔁 39 💬 3 📌 5#Cancer #Epigenomics
15.11.2024 17:29 — 👍 1 🔁 0 💬 0 📌 0
Pleasure to continue our collaboration with Marcus Wilson and group and play a small part in this study which is now out in the paper version. Read on to learn more about DNMT3A's interaction with H3K36me2 and H2AK119Ub modified nucleosomes:
www.embopress.org/doi/full/10....
#epigenetics
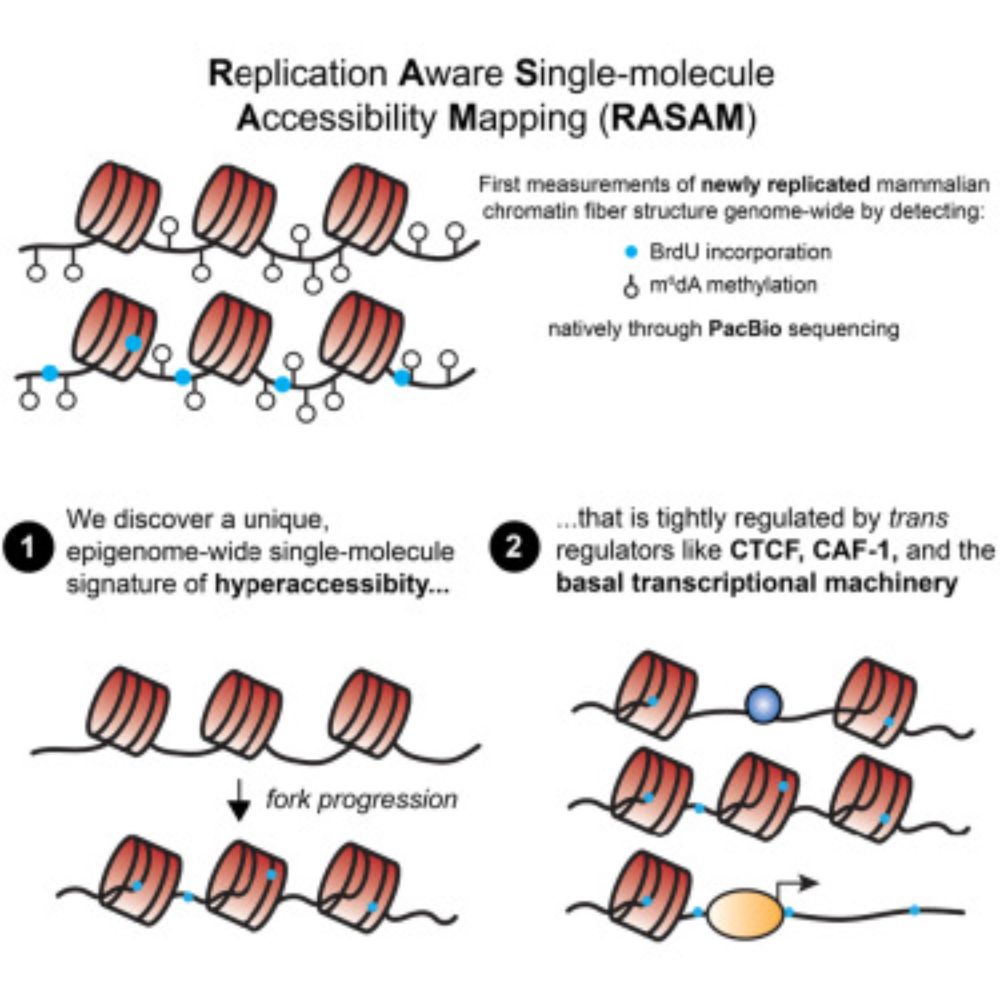
The latest from our group, led by Megan Ostrowski and @martyyang.bsky.social, is now published in final form (www.cell.com/cell/fulltex...! Many thanks to our excellent peer reviewers for suggesting several experiments (including CAF-1 perturbation) to really improve the study =) #epigenetics
15.11.2024 15:35 — 👍 207 🔁 75 💬 14 📌 9