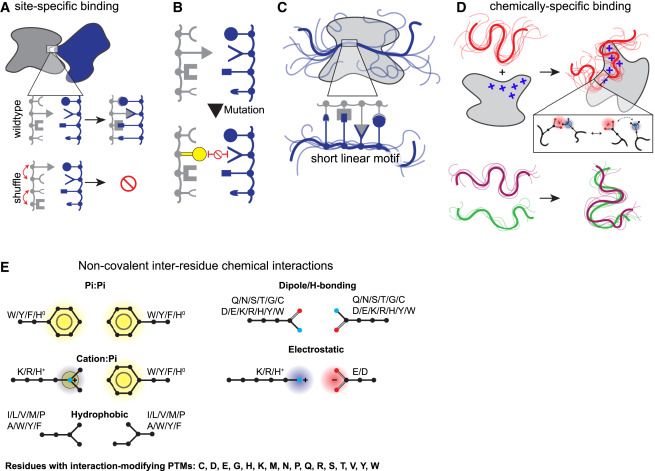
Nipah virus G protein with 16813 ligands (mostly waters) mapped from similar structures.
ChimeraX can run Foldseek to find similar structures, such as distantly related homologs, and analyze the results, for example, mapping all ligands onto your query structure. Here are ligands mapped onto Nipah virus G protein. www.rbvi.ucsf.edu/chimerax/dat...
06.03.2025 23:06 — 👍 96 🔁 36 💬 2 📌 5
Centromere evolution isn't a sudden switch!
Our study shows centromere transitions are a step-by-step process driven by a combination of drift and selection. Discover how the kinetochore interface shapes this gradual change in our new preprint 🥳 doi.org/10.1101/2025.01.16.633479 🧵(1/8)
17.01.2025 11:17 — 👍 103 🔁 40 💬 2 📌 5
🔥🔥🔥🔥🔥🔥
18.01.2025 02:27 — 👍 4 🔁 2 💬 0 📌 0
The more I think about it, the more convinced I am that the holy grail here is if this can go beyond predicting ratio of folded to unfolded, to also predicting ratio of folded state A to folded states B, C, D, etc. Example from doi.org/10.1016/j.molcel.2024.04.008 w/ changes in the transporter OCT1
11.12.2024 07:52 — 👍 39 🔁 6 💬 4 📌 0
It was fantastic and humbling to receive a personal email from @J_A_C_S, the editor-in-chief, congratulating us on this work. Malay_Moldal and others deserve all the credit.👏
10.12.2024 17:14 — 👍 4 🔁 1 💬 1 📌 0
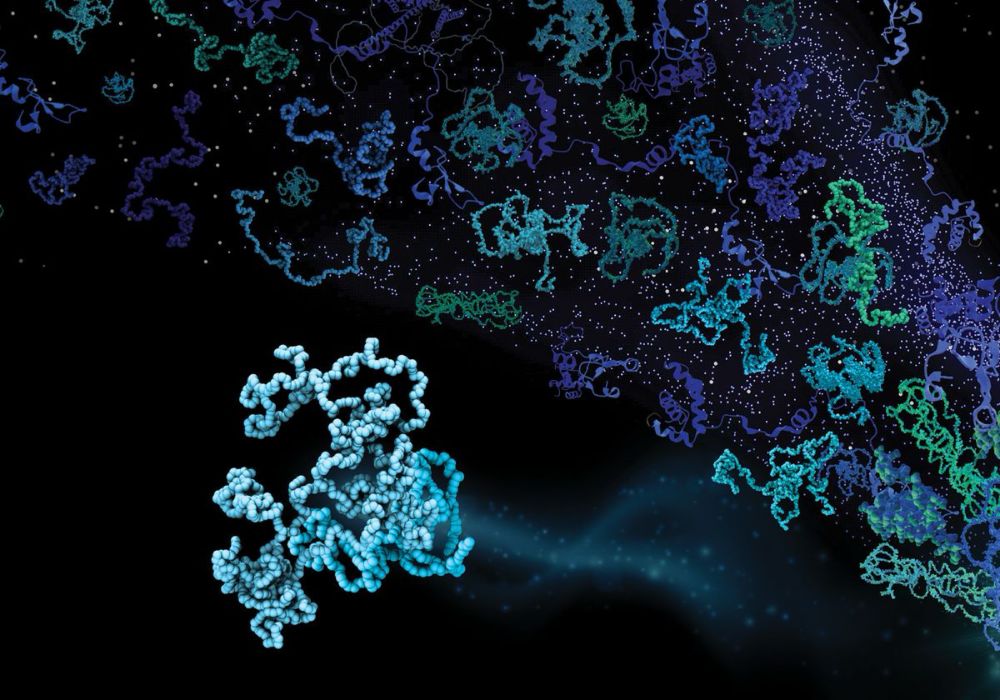
Meet the CALVADOS RNA model
Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪
29.11.2024 08:25 — 👍 89 🔁 21 💬 4 📌 2
This is pretty impressive--closing the gap between superresolution microscopy and structural biology is one of the Holy Grails and this is a big step in that direction--check out the nuclear pore imaging
29.11.2024 00:27 — 👍 53 🔁 12 💬 2 📌 0
The Dynamic Lives of Intrinsically Disordered Proteins
Shapeshifting proteins challenge a long-standing maxim in biology.
Very nice article by Danielle Gerhard in The Scientist on
“The Dynamic Lives of Intrinsically Disordered Proteins”
with quotes from Gabi Heller, @alexholehouse.bsky.social and myself about our shared love of and fascination with these proteins
www.the-scientist.com/the-dynamic-...
16.09.2024 17:02 — 👍 11 🔁 8 💬 0 📌 0
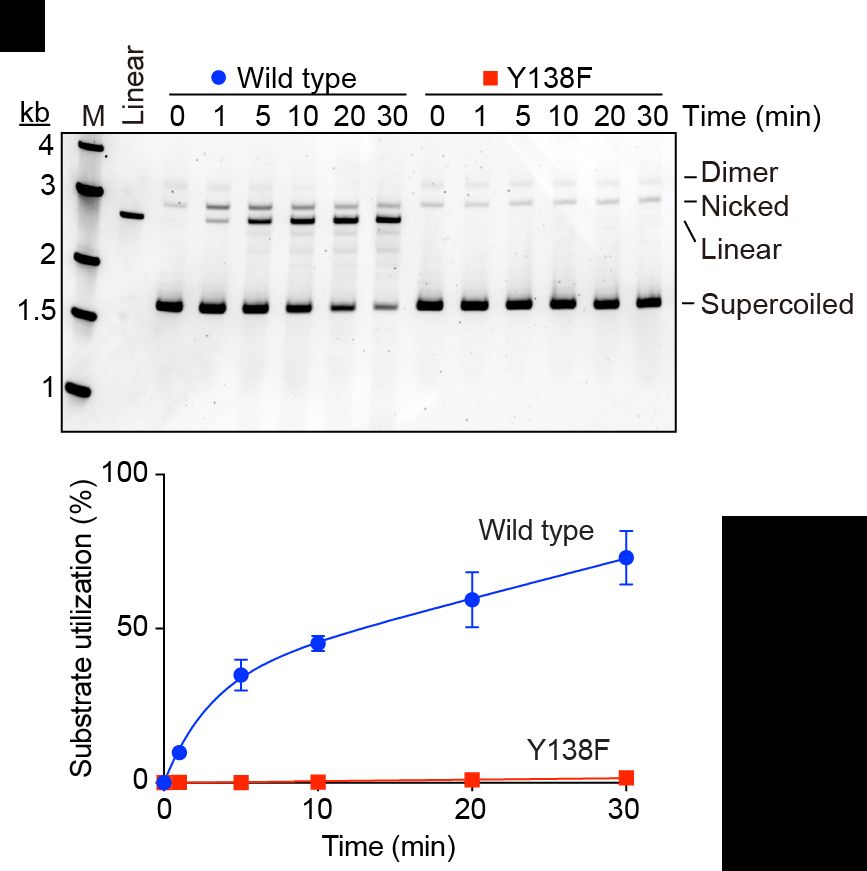
Experimental results showing DNA break formation by purified SPO11 protein. Above is an image of an electrophoresis gel, below is a graph showing quantification of the DNA cleaving activity
Hi Bluesky. For my first post, I'd like to advertise the latest preprint from my lab, describing at long last our reconstitution of SPO11-dependent double-strand break formation from purified recombinant proteins. Outstanding work from PhD student Zhi (Zack) Zheng.
23.11.2024 16:00 — 👍 156 🔁 51 💬 5 📌 5
Excited to share our new study with the Müller and @SchullerJm labs, online @nature: https://www.nature.com/articles/s41586-022-04971-z
This is T. kivui, an anaerobic bacterium. It uses hydrogen energy to store #CO2. How does it do it? #CryoET revealed membrane-anchored bundles of...
21.07.2022 04:51 — 👍 189 🔁 57 💬 8 📌 9
DPhil student in the Mommersteeg Group #cavefish #heartregeneration @UniofOxford | alum @ElitasLab @sabanciuni
I love/ teach chemistry 🐟🧪🫀
Senior Scientist • Arctic chronoecology • Avian migration. Leading terrestrial bird research and monitoring at Pinngortitaleriffik, GL.
http://nphuffeldt.wordpress.com
Personal account. Opinions & posts my own
Research group based at the University of Edinburgh investigating parasite evolution and ecology.
*Posts by various Reece lab members*
thereecelab.com
Chronobiologist and hibernation researcher based in the Arctic. Torpor arousal cycling. Photoperiodism. Circaunnual rhythms.
ASTI is an international centre of excellence in seasonal-timekeeping at UiT: The Arctic University of Norway.
BioClocks UK is a hub for biological rhythms research in the UK, supporting researchers to deliver impact through outreach, training, and community-specific resources.
Website: https://www.bioclocks.uk/
Find out more: https://doi.org/10.1098/rstb.2023.034
Marine biology & Chronobiology & Ecophysiology & Ecotoxicology
(She/her)
Developer of Wolfram Language data science pipelines for chronobiology researchers
Professor of #Biology at Universidade Federal da Fronteira Sul. I study #chronobiology and #sleep.
sites.google.com/site/felipebeijamini
She/her | PhD Student in the Oakley lab at UCSB, studying the evolution of biological rhythms in bioluminescent ostracods. | I like crunchy invertebrates and taking long walks on Caribbean beaches 🏖️
Microbiologist at UC Santa Cruz. Streptococcus pneumoniae, circadian rhythms, pneumonia, inflammation. www.kimmeylab.com
Assistant Professor at @WUSTLBio. Using genomics to study the intersection of Plant Defense, Circadian Rhythm and Development with genomics. I ❤️ dataviz
(he/him/his) BenMansfeldLab.com
Circadian neurobiologist at Texas A&M University
PI of the Jones Lab
jones-lab.org
Plant circadian biology at Department of Plant Sciences University of Cambridge.
Associate Professor in the School of BioSciences, University of Melbourne.
Metabolic signalling, circadian clocks.
Teacher of Genetics. Researcher of plants.
Dad. Runner.
https://blogs.unimelb.edu.au/haydonlab/
Head of plant genomics at Earlham Institute, #Circadian, #Genomics, #Bioinformatics, #wheat, #Arabidopsis. Note, my use of English is unconventional. Co-founder of TraitSeq. Programme lead for DECODING BIODIVERSITY https://tinyurl.com/4k8b8x2x
BBSRC Research Fellow @ the John Innes Centre, UK; from Montes Claros, Brazil; stares at clocks and bothers cyanobacteria for a living 🏳️🌈🇧🇷 ela/she/ella
Researcher studying adaptation of organisms to the Arctic light while trying to adapt to the Arctic light herself. ⏰🐦⬛🐌
Evolutionary ecologist working on how species adapt to their changing world. Lead PI of LTER-LIFE, an infrastructure to build Digital Twins of ecosystems. Head of the Animal Ecology departement at NIOO-KNAW and professor at Groningen University.