Congrats Rita, and thanks for everything you did at Nature Methods! Hopefully our paths cross again at some point 😀
06.08.2025 12:08 — 👍 1 🔁 0 💬 1 📌 0
SMLMS 2025 – Bonn, August 27th – 29th
Welcome to our blue space! SMLMS 2025 is actively in the making! Soon we will be able to reveal our amazing speaker line-up here in Bonn! Stay tuned! While waiting, maybe already bookmark smlms.org! 👈
See you in August 2025 in Bonn!
14.01.2025 13:05 — 👍 18 🔁 7 💬 0 📌 2
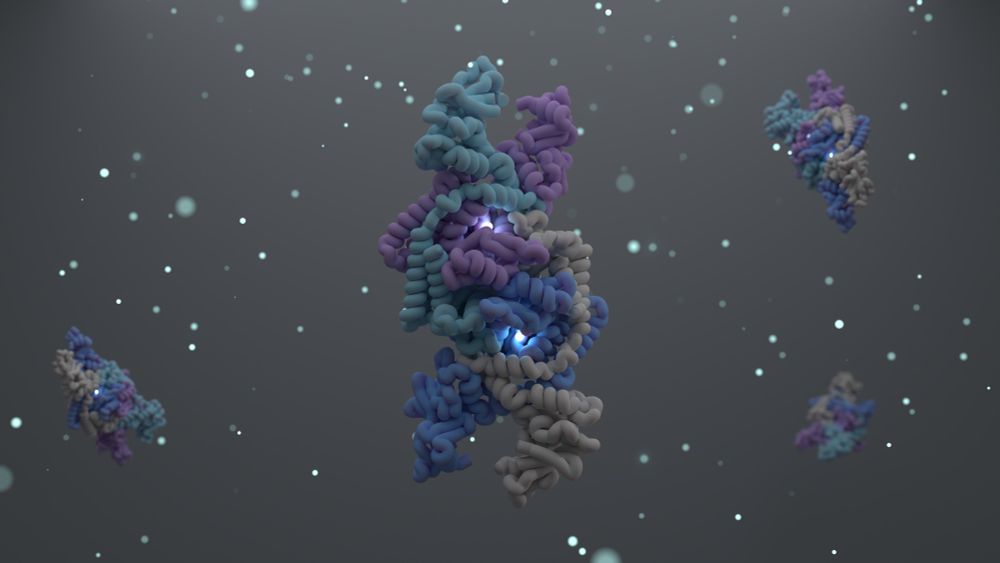
What a great way to end the year! ✨
Today in @cellpress.bsky.social we report the structure and function of the Shedu anti-phage defense system.
tinyurl.com/4crj6dnx
A long 🧵...
31.12.2024 15:38 — 👍 97 🔁 34 💬 6 📌 3

Razendsnel meerdere eiwitten tegelijk volgen in een cel: met deze nieuwe methode lukt het
Wetenschappers hebben een methode ontwikkeld waarmee voor het eerst razendsnel de bewegingen van meerdere eiwitdeeltjes in een bacteriële cel tegelijk gevol...
This week, I was interviewed by the Dutch newsradio
@bnrnieuwsradio.bsky.social about my latest work: TARDIS, a computationally novel way to track single proteins inside living cells. Listen to my radio-debut here (Dutch): bnr.nl/podcast/wete... or read all about the research here: rdcu.be/dv1sr
01.02.2024 15:53 — 👍 2 🔁 0 💬 0 📌 0
Openly accessible here: rdcu.be/dv1sr !
15.01.2024 11:10 — 👍 1 🔁 0 💬 0 📌 0
Finally, huge kudo's to all my labs in all three countries (WUR, NL, CarnegieMellon, USA, and UniBonn GER), and to funding from the #Humboldt Postdoc Fellowship @humboldt-foundation.de , Bonn Argelander program, VLAG@WUR, NSF, CarnegieMellon, and UniBonn! (8/8)
15.01.2024 10:25 — 👍 1 🔁 0 💬 0 📌 0
This method wouldn't be here without @uendesfelder.bsky.social , @hohlbeinlab.bsky.social and Dr. Turkowyd, and I love the improvements during the review process led by @ritastrack.bsky.social - it increased the fundamental, mathematical underpinning of TARDIS ánd allows more flexibility in fitting!
15.01.2024 10:23 — 👍 1 🔁 0 💬 1 📌 0

GitHub - kjamartens/TARDIS-public: Public releases of TARDIS
Public releases of TARDIS. Contribute to kjamartens/TARDIS-public development by creating an account on GitHub.
TARDIS promises to open up the possibility of performing spt data analysis in/with wildly novel conditions/probes. It will also directly benefit from all future endeavors in (mobile) particle localization at high density. Try TARDIS yourself here: github.com/kjamartens/T...
15.01.2024 10:21 — 👍 1 🔁 1 💬 1 📌 0

We used TARDIS to look at the in vivo movement of RNA polymerase, and show that we can easily lower the measurement time by a factor of ~5. This was limited by the required high-density localization, NOT by TARDIS performance. (5/8)
15.01.2024 10:21 — 👍 0 🔁 0 💬 1 📌 0
We went to crazy lengths to try to overwhelm TARDIS, but only managed to get a 'suboptimal' error of ~5% once we (far) surpassed the limits of mobile single-molecule localization AND had only 1.5 loc/traj on average AND > 50% of the dataset was pure noise (!) (4/8)
15.01.2024 10:21 — 👍 1 🔁 0 💬 1 📌 0

This concept accurately obtains either an analytically fitted distribution (anything is possible here), or a jump-distance histogram. It is far more robust than any tracking algorithm (at high complexity), and constantly surprised us with its robustness. (3/8)
15.01.2024 10:21 — 👍 0 🔁 0 💬 1 📌 0

TARDIS is a conceptually new method to perform single-particle tracking analysis: all localizations are compared to themselves with a time-shift. The intraparticle population is separated from the interparticle distribution by observing time delays longer than track lengths: (2/8)
15.01.2024 10:20 — 👍 3 🔁 2 💬 1 📌 0
TARDIS (Temporal Analysis of Relative Distances) is now out in
Nature Methods! nature.com/articles/s41... It redefines single-particle tracking: spt is no longer limited by tracking algorithms - TARDIS offers at least 10x higher throughput, exceptional noise robustness: a 🧵 (1/8)
15.01.2024 10:20 — 👍 12 🔁 2 💬 4 📌 0
Our group at LMU Munich and @mpibiochem.bsky.social uses DNA nanotechnology to develop next-generation super-resolution microscopy techniques. #DNAPAINT
Physicist | PhD Student @JungmannLab @MPI_Biochem @LMU_Muenchen
Super-resolution microscopy 🔬🧬 Spatial-omics technology 👨🎨🌈 Neuroscience Applications 🧠
Mathematician facing biomedical image processing challenges 💻🔬🧚♂️🦄
https://henriqueslab.org/pages/egdmariscal
@embo.bsky.social fellow at @henriqueslab.bsky.social @aiopticalbiolab.bsky.social
Former @arratemunoz.bsky.social @deniswirtz.bsky.social
Group leader of the Computational Microscopy for Cell Biology (CMCB) lab @ SciLifeLab, Stockholm.
Physicist studying cells with ML and fancy microscopes!
Assistant professor at IISc Bangalore. Interested in DNA nanotechnology, single-molecule biophysics, super-resolution imaging, chromatin organization.
Lab webpage: www.ganjilab.org
Single-molecule biophysics group at Goethe University Frankfurt/Germany. Super-resolution microscopy. Optical structural cell biology. Computational microscopy. Posts by MH.
http://www.smb.uni-frankfurt.de
#superresolution, #livecellimaging, #imageanalysis
🧠Neuroscientist specializing in 🔬 super-resolution microscopy
exploring #glycobiology 🍭 to broaden understanding of cellular processes.
Sharing insights on:
- our #research,
- notable studies by peers,
- and the intersection of 🎨 #art & 🧬 #science.
Musings about science, biophysics, inference, and occasionally Bach and Shostakovich
Group leader in Imaging Infection at the University of Bonn
https://www.ifmb.uni-bonn.de/en/research/rg-scherer/
Exploring frontiers of AI and its applications to science/engineering
Formerly: PhD/postdoc in Berkeley AI Research Lab + UC Berkeley Computational Imaging Lab
https://henrypinkard.github.io/
Professor of Physics (https://www.joerg-enderlein.de), Editor-in-Chief of Biophysical Reports (https://www.cell.com/biophysreports/home), Science & Arts, 🏳️🌈 | biophysics | single-molecule spectroscopy | super-resolution microscopy | nano-optics & plasmonics
CNRS Research director, Nanobio ISMO #superresolution #SMLM #FLIM #SAF #ModLoc #Microscopy @gdrimabio codirector. Scientific advisor for @abbelight. #Foodlover
Endurance sports aficionado - Skule and Gopher alum - always eager to learn and up for a chat over a coffee. Single molecule fan - from molecular crystals to protein complexes. Builder of tools and fan of all things imaging
Professor of physics and bioengineering @EPFL. Microscopy and mitochondria devotee. This view is all mine.
Microscopy builder, fluorescence enthusiast, let’s photo-switch
The Prevedel lab @EMBL Heidelberg. We develop high-speed, deep tissue as well as Brillouin microscopy techniques. More details @ www.prevedel.embl.de
Necessity is the mother of invention or "Not macht erfinderisch"
I'm a physicist; I invent techniques to measure and control biological systems.
Homepage: andrewgyork.github.io
Researcher in Optics and Biophysics @ EPFL | Fluorescence and Label-Free Single Molecule Imaging | Numerical Methods and Simulation
https://kylemdouglass.com
Microscopy core director MIT. Cell biology. Amateur musician: mandolin family and trombone. Cambridge MA





