
Now online! A non-catalytic role for RFC in PCNA-mediated processive DNA synthesis
29.01.2026 13:42 — 👍 3 🔁 2 💬 0 📌 0
Now online! A non-catalytic role for RFC in PCNA-mediated processive DNA synthesis
29.01.2026 13:42 — 👍 3 🔁 2 💬 0 📌 0
FoldMason is out now in @science.org. It generates accurate multiple structure alignments for thousands of protein structures in seconds. Great work by Cameron L. M. Gilchrist and @milot.bsky.social.
📄 www.science.org/doi/10.1126/...
🌐 search.foldseek.com/foldmason
💾 github.com/steineggerla...

Pleased to share our recent article in PNAS - a collaboration with @jessicaandreani.bsky.social & Pablo Radicella, with important roles played by many members of each team.
A tripartite protein complex promotes DNA transport during natural transformation in Firmicutes www.pnas.org/doi/10.1073/...
New paper alert 🚨
I am super happy about it, for many reasons!
👥 It was a great collaborative project with Calum @chgjohnston.bsky.social and J. Pablo Radicella, designed a long time ago (in the pre-AlphaFold era - can you imagine?) together with @polardlab.bsky.social and @raphguerois.bsky.social

How do new centromeres evolve while staying compatible with the division machinery?
Discover it in our new Nature paper! We show centromeres transition gradually via a mix of drift, selection, and sex, reaching new states that still work with the kinetochore.
👉 doi.org/10.1038/s41586-025-09779-1
Structural basis for fork reversal and RAD51 regulation by the SCF ubiquitin ligase complex of F-box helicase 1 pubmed.ncbi.nlm.nih.gov/41256537/ #cryoEM
21.11.2025 01:10 — 👍 2 🔁 2 💬 0 📌 0
Thrilled to share that the final piece of my PhD work is now on bioRxiv! biorxiv.org/content/10.1... With support from @nvidia and the @NSF, we used AlphaFold to screen 1.6M+ protein pairs, revealing thousands of potential novel PPIs. All data can be viewed at predictomes.org/hp
12.11.2025 21:26 — 👍 163 🔁 67 💬 5 📌 4
www.nature.com/articles/s42...
Latest from the lab. The conclusion is in the title!
Basically, we found that the KMN complex (outer kinetochore) is fully conserved between plants and fungi/animals, showing deep origin. (reminder, you are closer to a mushroom than a mushroom is to a plant.)

Among the anti-recombinases, FIGNL1 rules them all. So much that inactivating it brings BRCA2-deficient cells to life. Who is responsible for RAD51 loading without BRCA2/FIGNL1, check out the paper to find out! Great collaboration with @raychaudhurilab.bsky.social
www.science.org/doi/10.1126/...

Excited to release BoltzGen which brings SOTA folding performance to binder design! The best part of this project is collaborating with a broad network of leading wetlabs that test BoltzGen at an unprecedented scale, showing success on many novel targets and pushing the model to its limits!
26.10.2025 22:40 — 👍 103 🔁 41 💬 3 📌 5
Prime editing sensor libraries evaluate diverse genetic variants in their endogenous genomic contexts go.nature.com/49Q1WCQ
rdcu.be/eLVts


Super excited about first Shendure/Baker Lab collaboration & preprint on a multiplex sequencing-based strategy for screening de novo proteome editors in mammalian cells. Kudos to the brilliant Chase Suiter (not here) & @greenahn.bsky.social on the work! Preprint here:
www.biorxiv.org/content/10.1...
Thrilled to announce our new preprint, “Protein Hunter: Exploiting Structure Hallucination within Diffusion for Protein Design,” in collaboration with @Griffin, @GBhardwaj8 and @sokrypton.org
🧬Code and notebooks will be released by the end of this week.
🎧Golden- Kpop Demon Hunters
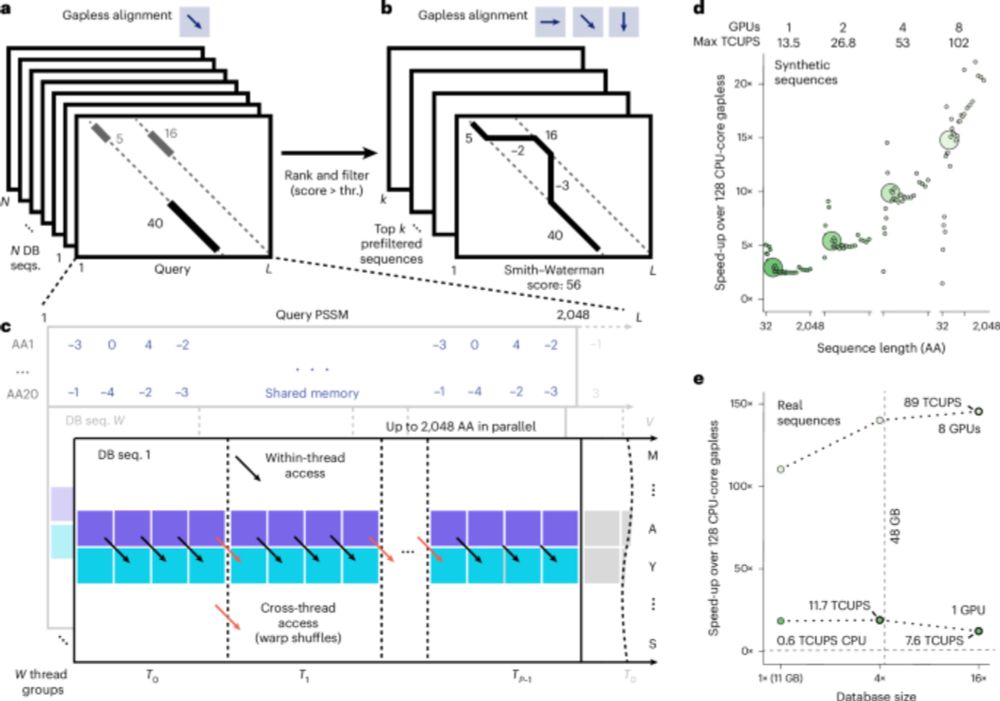
GPU-accelerated MMseqs2 offers tremendous speedup for homology retrieval, protein structure prediction with ColabFold, and protein structure search with Foldseek. @martinsteinegger.bsky.social @milot.bsky.social @machine.learning.bio
www.nature.com/articles/s41...
Exciting to see our protein binder design pipeline BindCraft published in its final form in @Nature ! This has been an amazing collaborative effort with Lennart, Christian, @sokrypton.org, Bruno and many other amazing lab members and collaborators.
www.nature.com/articles/s41...
📌
05.08.2025 21:10 — 👍 0 🔁 0 💬 0 📌 0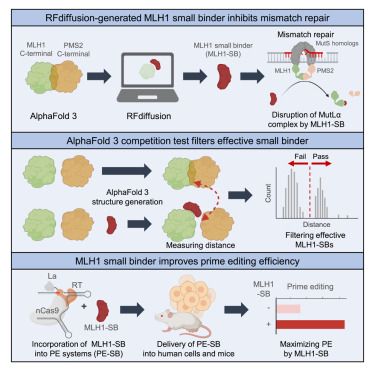
Now online! AI-generated MLH1 small binder improves prime editing efficiency
05.08.2025 18:52 — 👍 1 🔁 2 💬 1 📌 0
Folddisco finds similar (dis)continuous 3D motifs in large protein structure databases. Its efficient index enables fast uncharacterized active site annotation, protein conformational state analysis and PPI interface comparison. 1/9🧶🧬
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
AI just cracked de novo antibody design with a 100x leap! 🧪
Chai-2, a next-gen multimodal generative model, is shaking up protein design. With a 16% hit rate in fully de novo antibody design—over 100 times better than previous methods—it’s making drug discovery faster and more precise than ever.
Excited to unveil Boltz-2, our new model capable not only of predicting structures but also binding affinities! Boltz-2 is the first AI model to approach the performance of FEP simulations while being more than 1000x faster! All open-sourced under MIT license! A thread… 🤗🚀
06.06.2025 13:46 — 👍 215 🔁 91 💬 10 📌 11
🚀 Excited to release BoltzDesign1!
✨ Now with LogMD-based trajectory visualization.
🔗 Demo: rcsb.ai/ff9c2b1ee8
Feedback & collabs welcome! 🙌
🔗: GitHub: github.com/yehlincho/Bo...
🔗: Colab: colab.research.google.com/github/yehli...
@sokrypton.org @martinpacesa.bsky.social
@science.org 🧫🧬❄️🔬 Molecular basis of influenza ribonucleoprotein complex assembly and processive RNA synthesis | Science www.science.org/doi/10.1126/...
@yiweichang.bsky.social www.yiweichanglab.org @jiwasa.bsky.social #virology #Influenza #Cryo-EM #StructuralBiology #RNA #polymerase

Forget liquid-liquid phase separation, here comes solid phase transition 😱
www.nature.com/articles/s41...

Pleased to share a new preprint - a great collaboration with @jessicaandreani.bsky.social and the Guerois lab at I2BC, and the Radicella lab at the CEA. Using stuctural modelling, we reveal & validate a 3 protein complex involved in DNA import during transformation.
www.biorxiv.org/content/10.1...

Please see our latest paper on the role of EXO1 in meiosis: "EXO1 promotes the meiotic MLH1-MLH3 endonuclease through conserved interactions with MLH1, MSH4 and DNA". Congratulations to both first authors, Megha Roy and Aurore Sanchez and thanks to all our collaborators!
05.05.2025 07:22 — 👍 44 🔁 11 💬 0 📌 0
Happy to share our DNA Repair special issue on the replication stress response curated with @vindignilab.bsky.social.
www.sciencedirect.com/special-issu...
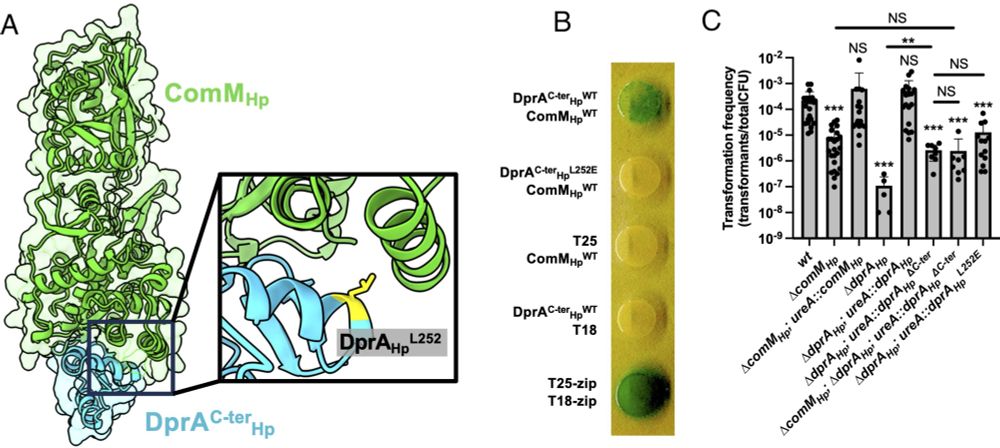
📣 Happy to announce that "DprA recruits ComM to facilitate recombination during natural transformation in Gram-negative bacteria" just got published in PNAS www.pnas.org/doi/abs/10.1...
🧬🧶👩🔬👩💻 Collaborative and interdisciplinary work, together with the groups of Ankur Dalia and J. Pablo Radicella
🧬 @science.org Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis | Science www.science.org/doi/10.1126/...
14.04.2025 05:04 — 👍 13 🔁 2 💬 1 📌 0I am a professor at Columbia University. All of the student NIH training grants have been canceled and now there are reports that ALL funding will be frozen. Why are Universities not banding together and speaking out publicly and forcibly about governmental attacks on biomedical research?
11.04.2025 16:53 — 👍 1309 🔁 408 💬 53 📌 19