Out now!
09.02.2026 19:20 — 👍 2 🔁 0 💬 0 📌 0Out now!
09.02.2026 19:20 — 👍 2 🔁 0 💬 0 📌 0
Second up this Wednesday, Jiayuan Cui will tell us about her exciting and rigorous research biochemically characterizing a radical SAM enzyme involved in archaeal monolayer lipid (GDGT) cross-linking. Check your email for the Zoom link or visit our website (link in bio) for more info on how to join!
15.12.2025 15:22 — 👍 5 🔁 3 💬 1 📌 0
Archaea Power Hour is back for our final session of the semester this WEDNESDAY, Dec. 17th, at 10AM EST / 4PM CET. We’ll start off with an intriguing talk from @fmacleod.bsky.social about his work uncovering unique intracellular membrane-bound compartments in an Asgard archaeon!!!
15.12.2025 15:11 — 👍 6 🔁 3 💬 1 📌 0“Happy Holidarchaeays” to you too
15.12.2025 01:39 — 👍 2 🔁 1 💬 0 📌 0Sure, we have plenty beautiful examples to share! Who should I contact?
09.12.2025 22:39 — 👍 1 🔁 0 💬 1 📌 0Lastly, for those at #CellBio25, we will be having a social TONIGHT at Little Gay Pub (102 S 13th Street) at 8 p.m.! Come meet the LGBTQ+ committee and let's celebrate another great conference! #outinstem #LGBTQIA #LGBTQ #queerinstem #cellbio #cellbiology #ascb #CellBio2025
09.12.2025 13:18 — 👍 5 🔁 5 💬 0 📌 0Tomorrow!! Special Pohlschröder lab double feature, because we halove their work (going to pun hell now)
26.11.2025 03:20 — 👍 1 🔁 0 💬 0 📌 0
Archaeasalutations!! Archaea Power Hour is back tomorrow (Wednesday) at 10 AM EST / 4 PM CET!! Check your email for the Zoom link or sign up to join our mailing list here: docs.google.com/forms/d/e/1FAI…. Check out the two exciting talks we have planned for you:
25.11.2025 20:01 — 👍 9 🔁 4 💬 1 📌 2Stunning! Congrats @buzzbaum.bsky.social et al!
07.11.2025 23:10 — 👍 4 🔁 0 💬 1 📌 0Rest in power Zara, you’ll be missed
07.11.2025 15:31 — 👍 1 🔁 0 💬 0 📌 0
Glad to share our paper out today @NatureEcoEvo: “Serial innovations by Asgard archaea shaped the DNA replication machinery of the early eukaryotic ancestor”. www.nature.com/articles/s41... #microsky #archaeasky
21.10.2025 15:05 — 👍 64 🔁 28 💬 5 📌 4Don’t miss the first APH meeting of the season this coming Wednesday!
17.10.2025 17:06 — 👍 3 🔁 2 💬 0 📌 0
Welcome to our first postdoc in the ACO Lab, Andy Garcia! 🎉
15.09.2025 20:09 — 👍 17 🔁 0 💬 0 📌 0
Supporting each other as #newPI is very important, and @antonioserapio.bsky.social is great at it
01.09.2025 22:27 — 👍 0 🔁 0 💬 0 📌 0
🚨 🚨 🗓️ The deadline to submit your abstract for the #ASCB2025 meeting in Philadelphia is this Wednesday, September 3!!
Dyche Mullins and I are hosting the session “Cell Biology at the Extremes”. Thermophiles, psychrophiles, tardigrades, giant bacteria… now is your time to shine!! 🚨
New hot fluorescent tag dropped!
25.06.2025 18:04 — 👍 5 🔁 0 💬 0 📌 0Thank you!
30.05.2025 12:47 — 👍 1 🔁 0 💬 0 📌 0
We’ve uncovered Asgard chromatin structures formed by a Hodarchaeal histone : closed hypernucleosome conserved in archaea and an open form resembling the H3-H4 eukaryotic octasome. Fantastic work by @harshranawat.bsky.social!
www.biorxiv.org/content/10.1...
#Chromatin #Asgard #Archaea #cryoEM
Thanks!
29.05.2025 18:51 — 👍 0 🔁 0 💬 0 📌 0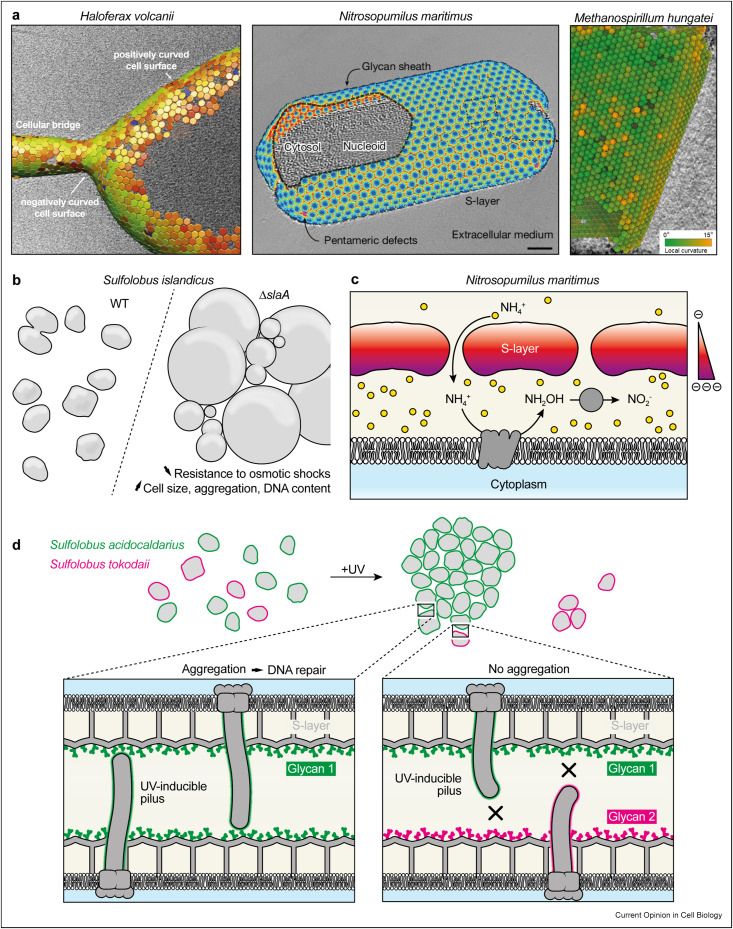
Archaeal S-layer proteins can adopt different symmetries that can locally accommodate areas of high curvature. The S-layer is also involved in the control of turgor and osmotic pressure, ion transport, cell-cell interactions and mating.
29.05.2025 16:45 — 👍 22 🔁 6 💬 1 📌 2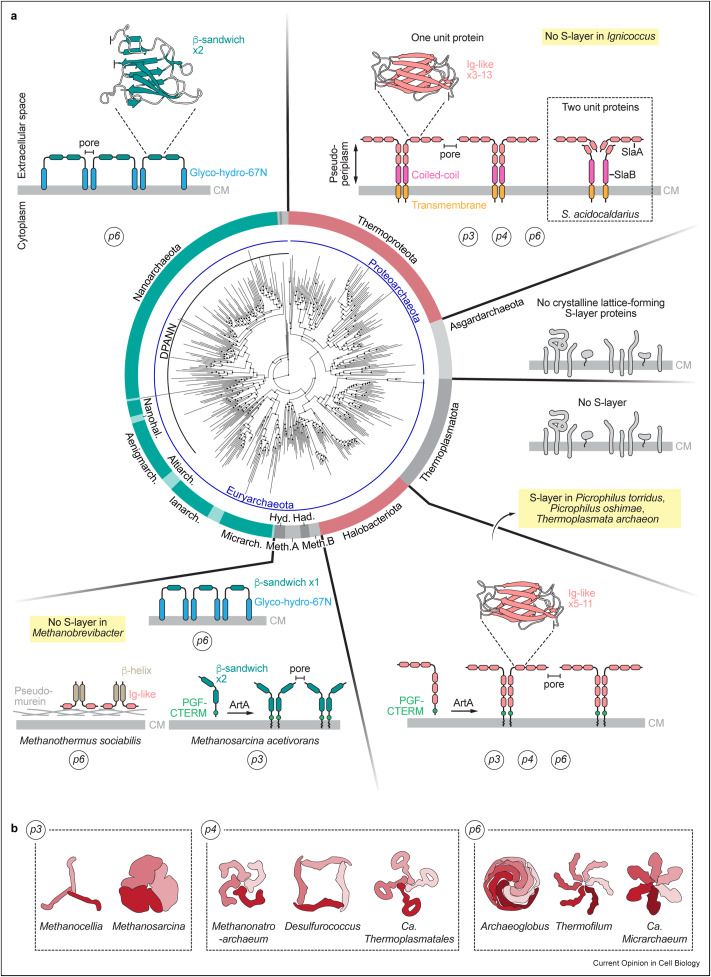
The S-layer is the most widespread and fundamental component of archaeal cell envelopes and may represent one of the earliest and most fundamental cell wall polymers found in microorganisms
29.05.2025 16:43 — 👍 12 🔁 7 💬 0 📌 0
The archaeal S-layer is not only pretty, it also carries crucial cellular functions, and the list keeps growing! @sshamphavi.bsky.social @marleenvw.bsky.social @archaellum.bsky.social and I wrote a little sheath sheet (+10 pts if you got the pun)
📖 Curr Opin Cell Biol
🔗 doi.org/10.1016/j.ce...
What a year for the cell biology of archaeal chromosome segregation! 🧬 @joeparham19.bsky.social and @buzzbaum.bsky.social now add pieces to the SegAB puzzle by connecting it with cell division
29.05.2025 15:55 — 👍 8 🔁 3 💬 0 📌 0
To explore how ring assembly and DNA segregation might be coupled, we turned our attention to the ParA homologue, SegA, and its partner SegB, using antibodies kindly gifted by @acharlesorszag.bsky.social (see www.biorxiv.org/content/10.1...)
29.05.2025 15:36 — 👍 3 🔁 1 💬 1 📌 0
In just 2 days on WEDNESDAY, May 14th at 10 AM EST / 4 PM CET, @fabaiwu.bsky.social will kick off our last spring APH session by telling us about his new research investigating the evolution of DNA replication machineries from Archaea to Eukaryotes!!! Visit our website (link in bio) for more info!
12.05.2025 21:39 — 👍 8 🔁 6 💬 1 📌 0
Think you're under pressure? Marion Illartin will close out our last APH session of the semester by telling us about her multidisciplinary approach to characterize how pressure regulates the SurR transcription factor in everyone's favorite barophile, Thermococcus barophilus!! Zoom in on May 14th!
12.05.2025 21:55 — 👍 2 🔁 3 💬 0 📌 0Don’t miss it!! Last APH of the season 🫶🏼
08.05.2025 17:21 — 👍 2 🔁 0 💬 0 📌 0Oop!
08.05.2025 14:01 — 👍 0 🔁 0 💬 0 📌 0If I may weigh in here: I avoid overfixing the cells, especially if using glutaraldehyde that can induce significant artefacts. I usually do 30-60 min at RT and rinse abundantly in buffer. Fixed cells in buffer can then be kept for days in the fridge :)
07.05.2025 19:51 — 👍 3 🔁 0 💬 0 📌 0Hahaha awww 🫶🏼 I’m very ignorant when it comes to anaerobic cultures. What are the challenging steps pre-fixation? The spinning?
07.05.2025 19:46 — 👍 0 🔁 0 💬 1 📌 0