Now out at #jbiolchem www.sciencedirect.com/science/arti...
28.05.2025 14:52 — 👍 7 🔁 3 💬 0 📌 0


Our full-day mini AI hackathon on genomic language models starts soon: 8.30am-8.30pm! With a whole team, we aim to test how gLMs work for two tasks:
1. SNP2GEX: personal variants to gene expression (unseen individuals & genes);
2. Seq2CellxTF: short regulatory to TF binding (unseen cell & TFs).
02.04.2025 00:22 — 👍 5 🔁 2 💬 1 📌 1
A case where the oodles of public lung cell line data generated as a result of COVID can be put to good use in cancer! Well done to Sojung Lee (PhD student), for the massive effort in data mining. (9/n)
17.03.2025 06:36 — 👍 2 🔁 0 💬 0 📌 0
Given that ERVK-7 expression is a potential biomarker for ICB response in LUAD, ERVK-7.long may be a more specific and easier to quantify. Controlling the epigenetic regulation of ERVK-7.long may also alter the immune microenvironment of LUAD. (8/n)
17.03.2025 06:36 — 👍 0 🔁 0 💬 1 📌 0

In addition to cell type specificity, the promoter of ERVK-7.long is driven by NF-κB, likely via TNF-α, while ERVK-7.short is already known to be regulated by IRFs. (7/n)
17.03.2025 06:36 — 👍 0 🔁 0 💬 1 📌 0


The promoter of ERVK-7.long was found to be lineage-specific, with H3K4me3 deposition in LUAD cells. ScRNA-seq confirmed that ERVK-7.long expression is restricted largely to LUAD cells, fibroblasts, and AT-2 cells, which may be the cell type of origin of LUAD cells. (6/n)
17.03.2025 06:36 — 👍 0 🔁 0 💬 1 📌 0
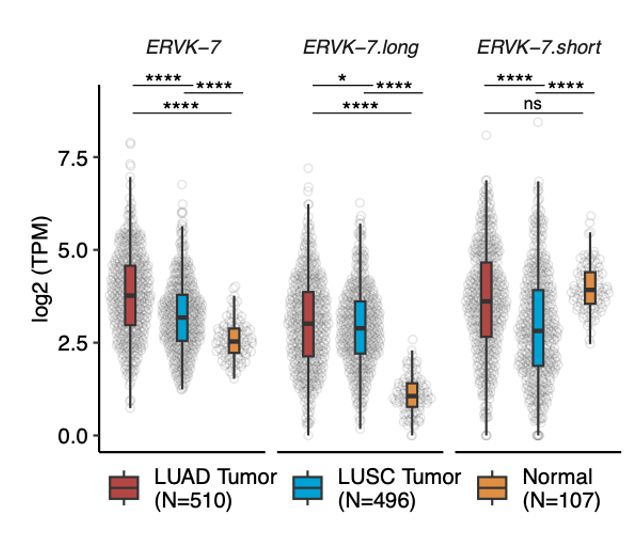
ERVK-7.long is highly expressed in LUAD, contributing significantly to the overall expression of ERVK-7. This suggests that the epigenetic regulation of ERVK-7.long may be key to ERVK-7 overexpression in patients. (5/n)
17.03.2025 06:36 — 👍 0 🔁 0 💬 1 📌 0

A key motivation for us is to understand the epigenetic regulation of ERVK-7 as it is frequently over-expressed but only amplified in ~10% of LUAD patients. (3/n)
17.03.2025 06:36 — 👍 0 🔁 0 💬 1 📌 0
https://www.nature.com/articles/s41586-023-05771-9
HERVs are generally silent or dysfunctional in the human genome. Yet recent studies have shown that the expression of certain copies, such as ERVK-7 (HERV-K102), is associated with autoantibodies and immunotherapy response in lung cancer patients t.co/21yyHijgMd. (2/n)
17.03.2025 06:36 — 👍 0 🔁 0 💬 1 📌 0
First post on Bluesky 🦋. Happy to share some new work on the epigenetic regulation of a human endogenous retrovirus (HERV) in cancer. @biorxivpreprint.bsky.social bsky.app/profile/bior... 🧵 (1/n)
17.03.2025 06:36 — 👍 1 🔁 0 💬 1 📌 2
McGill-Kyoto Joint PhD Student in Genomic Medicine 🧬 💻
Supervised by Dr. Guillaume Bourque @ McGill University🇨🇦 and Dr. Mitinori Saitou @ Kyoto University🇯🇵.
ASBMB drives discovery forward in academic, industry and governmental sectors, helping to generate new scientific knowledge today that fuels tomorrow’s biotechnological and medical breakthroughs.
www.asbmb.org | #ASBMB26
The American Society for Biochemistry and Molecular Biology advances scientific knowledge by reducing barriers to research. ASBMB journals, JBC, MCP & JLR, are gold open access.
www.asbmb.org/journals-news | #ASBMBJournals
PhD student | Davidson Lab | @wehi-research.bsky.social @unimelb.bsky.social | bioinformatics 🧬
Professor of Cancer Evolution. UCL Cancer Institute. Interested in cancer genomics, bioinformatics, and somatic evolution.
https://www.ucl.ac.uk/medical-sciences/divisions/cancer/our-research/cancer-genome-evolution
Group leader, Liccardi lab, Institute of biochemistry, University of Cologne.
We are interested in cell death inflammation and immunity.
Genomics, bioinformatics; attempting to say useful things about cancer and evolution.
Institute of Genetics and Cancer, University of Edinburgh
https://institute-genetics-cancer.ed.ac.uk
https://semplelab.com
Postdoctoral bioinformatician at the University of Edinburgh Institute of Genetics and Cancer.
Currently working on chromosomal instability and mutational landscapes in tumours.
Happiest when out in nature!
Postdoc in the Boulton Lab at the Francis Crick Institute. Currently working on dna damage.
PMGC offers high-quality, cost-effective genomics services to academic & commercial clients worldwide. Specializing in DNA seq, epigenomics, panel-based, single-cell & spatial genomics. Get in touch for custom unique solutions for your research!
PhD student studying colorectal cancer metastasis at the CRUK Scotland Institute
Arts and crafts nerd
Aging and cancer stem cell heterogeneity - ICREA research professor - Quantitative Stem Cell Dynamics lab
at IRB Barcelona 🇦🇷🇪🇺🇺🇸 fraticellilab.com
Computational biologist working on infectious disease
The Tumor Genomics Group (Aaltonen Lab) focuses on human tumor susceptibility as well as somatic cancer genomics.
Postdoc Fellow @ Uni Neuchâtel 🇨🇭 | Transposable Elements + SV, Evolution across scales | #openscience | #FirstGen | 🇬🇧🇫🇷 | Ski | Bike | Climb | Join TEsky: https://bsky.app/profile/did:plc:mw4y54k2y45j5dqxrbelwvmu/feed/aaad7mmgzkfuu
tobybaril.github.io
Prof in Human Genetics at McGill, Director of Bioinformatics at the McGill Genome Center and Director of @C3Genomics
https://computationalgenomics.ca/BourqueLab/
Interested in DNA damage, mutations, adaptation and evolution.
DKTK, DKFZ, LMU https://dktk.dkfz.de/en/research/dktk-researchers/anderson-group
Munich, Germany
Cancer researcher at Hunter College of the City University of New York & Weill Cornell Medicine. Views are my own.