25 years of the Chemistry Development Kit
Twenty five years ago the Chemistry Development Kit (CDK) was founded. The Chemistry and Internet (ChemInt2000) had just ended (it ran from 23 to 26 September) and my friend and I had taken the Amtrak night train from Washington to South Bend. At that time there were two leading Java applets for chemistry, JChemPaint and Jmol. I had hacked Chemical Markup Language support into both of them, and Dan Gezelter (Jmol and openscience.org), Christoph Steinbeck (JChemPaint), and me took the opportunity of being in North America to discuss if we could use a common code base. Chris’ _compchem_ had done something similar. Peter Murray-Rust, who had also attended ChemInt2000 like me and Chris did not attend.
I do not remember exactly, but I guess we must have met on the 28th and 29th? Maybe already on Wednesday. During this meeting we discussed a common data model (yes, Jmol used the CDK data model at some point) and somewhere during the meeting we wrote down a name for the project. There was the Java Development Kit, so this could be the Chemistry Development Kit. The name stuck.
A quick post like this cannot do credit to the history of the CDK, nor of everyone involved in the past or still is. You can browse some of the history of the CDK in my blog and in Chris’ blog. It has been an amazing journey and with a small grant just behind us (with Alyanne de Haan, René van der Ploeg, and Marc Teunis from Hogeschool Utrecht), and all the awesome things ongoing (new JChemPaint, various extensions, upgraded downstream tools), the CDK is alive and kicking.
A huge congrats and thanks to everyone (and every company and organization) who contributed code to the CDK with this huge milestone. There are a few people that I want to particularly thank (see the AUTHORS file for all names): Chris, who in the late nineties made a difference with open source in chemistry, Dan, for Jmol and hosting this memorable meeting at Notre Dame University, Rajarshi Guha, who operated _CDK Nightly_ for many years, well before Travis and Google Actions, Stefan, Miguel, Gilleain, and Christian, for many years of contributions to the CDK, and John Mayfield, the current CDK release manager.
25 years Chemistry Development Kit (CDK) and going strong! :) https://doi.org/10.59350/4ce2c-fxh02 https://chem-bla-ics.linkedchemistry.info/2025/09/28/25-years-of-the-chemistry-development-kit.html
Replying to this post will make it show up in my blog.
#openscience #chemistry #opensource
28.09.2025 08:41 — 👍 3 🔁 8 💬 0 📌 0
this week, 25 years ago, @steinbeck, @egonw, and @gezelter came together in South Bend., USA and founded the Chemistry Development Kit
#chemistry #openscience
26.09.2025 10:30 — 👍 0 🔁 8 💬 1 📌 0
Packages from the chemfp project
@dalke released chemfp 5.0, a Python package for cheminformatics fingerprint generation, search, and analysis, with support for CDK fingerprints
You can install it on Linux-based OSes using:
python -m pip install chemfp -i https://chemfp.com/packages/
#chemistry #python
24.09.2025 14:17 — 👍 0 🔁 2 💬 0 📌 0

Screenshot of a frame of one of the videos in the linked pull request, showing a chemical structure with various R-groups on the left (code and R group values), and on the right a list of SMILES combinatorial structures generated from the input on the left.
nice new R-group SMILES enumeration feature in the upcoming JChemPaint release, see https://github.com/JChemPaint/jchempaint/pull/225
#openscience #chemistry
11.09.2025 11:40 — 👍 0 🔁 7 💬 0 📌 0
Original post on fosstodon.org
pybacting 0.2.15 is released with Bacting 1.0.7 (see also https://social.edu.nl/@egonw/115040533430981814): https://pypi.org/project/pybacting/0.2.15/
pybacting is a Python wrapper around Bacting which provides functionality from the Chemistry Development Kit, OPSIN, Oscar4, BridgeDb, InChI […]
23.08.2025 10:51 — 👍 0 🔁 2 💬 0 📌 0

Screenshot of the book and the section "Exact Search" showing two scripts about exact structure matching. There is general text, but also two snippets of Groovy code, with links to show the full code example. At the bottom we see the 2D depictions of two chemical structures, isobutane and cycloproane
porting more content of the old Groovy #Cheminformatics with the #Chemistry Development Kit book to Markdown, here about substructure searching: https://cdk.github.io/cdkbook/substructure.html
14.08.2025 12:50 — 👍 1 🔁 4 💬 0 📌 0
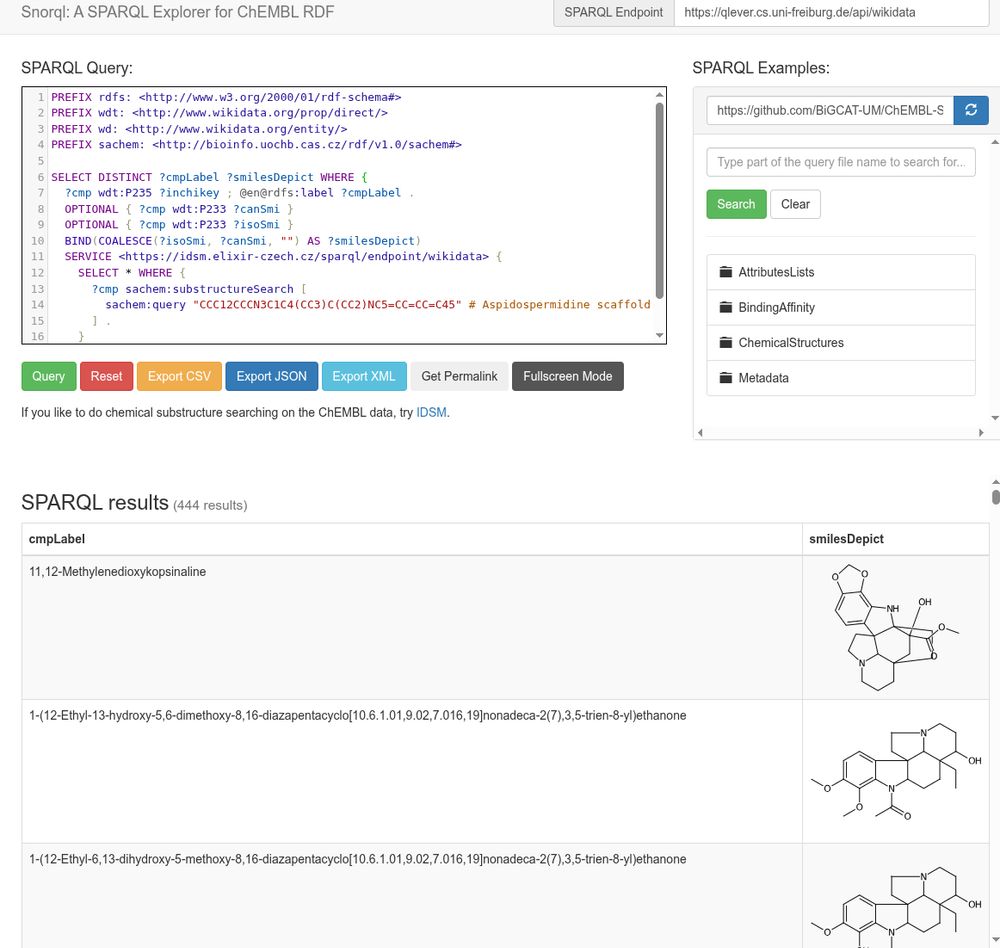
Screenshot of Ammar's Snorql UI, here wrapped around the Wikidata SPARQL instance by the QLever team, federating with the IDSM SPARQL endpoint for substructure searching, and using CDK Depict to convert the SMILES into 2D depictions in the results tables
oolonek reminded me how cool IDSM is (see https://doi.org/10.1186/s13321-021-00515-1) and works wonderfully in the rich chemistry SPARQL world (see https://doi.org/10.1186/1758-2946-3-15).
Here I mashed up @wikidata, IDSM, QLever, @cdk Depict, and Ammar […]
[Original post on social.edu.nl]
29.05.2025 10:16 — 👍 0 🔁 6 💬 0 📌 0

Screenshot of the link OpenAlex page, showing a query asking for all articles citing 5 articles (3 main articles, and 2 derivaties) and then the 4 most cited articles (citing the CDK) on the left, and some statistics on the right. These include the total citation sum (1,716), that 58.7% of the citing articles have been published open access, that they are mostly artices (1267), followed by preprints, reviews, book chapters, and dissertations (32). And that most citations come from the USA (380), followed by Germany (272), China (217), UK (210), and Switzerland (100).
Impact of our articles (and part of that of our source code; not every citation is an article where the CDK is used), according to OpenAlex. About 75 citations per year, totalling to 1716. And note how often those articles are cited.
20.04.2025 07:24 — 👍 0 🔁 0 💬 0 📌 0
Original post on fosstodon.org
#openscience #cheminformatics dates back to the late nineties with the emerging collaborative development of JChemPaint, Jmol, and the Chemical Markup Language. Sketch of the history by Chris Steinbeck: "The evolution of open science in cheminformatics: a journey from closed systems to […]
05.04.2025 04:30 — 👍 0 🔁 4 💬 1 📌 0
cdk/cdk: CDK 2.9
Interim release fixing an issue in CDK 2.10.
John release CDK 2.11, a quick bug fix release: https://doi.org/10.5281/zenodo.15105090 https://github.com/cdk/cdk/releases/tag/cdk-2.11
30.03.2025 09:52 — 👍 0 🔁 3 💬 0 📌 0
Original post on fosstodon.org
In "The Six Ds of Exponentials and drug discovery: A path toward reversing Eroom’s law" https://doi.org/10.1016/j.drudis.2025.104341
"however, software such as PaDEL and the Chemistry Development Kit (CDK) have emerged rapidly as free and powerful alternatives. Nowadays, many groups in both […]
26.03.2025 05:26 — 👍 0 🔁 6 💬 0 📌 0
Original post on fosstodon.org
Jonas Schaub: "Last week, I presented my work on algorithmic substructure extraction (scaffolds, functional groups, and aglycones) at the Chemistry Development Kit User Group Meeting (#CDK25UGM) in Maastricht.
You can now find my slides on Zenodo: https://doi.org/10.5281/zenodo.15058008" […]
22.03.2025 07:12 — 👍 0 🔁 5 💬 0 📌 0
![Screenshot of a record of Wikidata of a (P31 = instance of) "functional group", showing a regular SMILES with a "[*]" and a CXSMILES with the same but the "CX" addition "$_AP1$" causing the CDK-depiction to show a wigly line perpendicular to the bond "attaching the functional group" to the rest of the molecule.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:qj43mvyjyvzikd45py7f3q7s/bafkreib2h2h5lkocnyu2jtbdhvnksdy7djiry227lnr7uev7bjrelefpdm@jpeg)
Screenshot of a record of Wikidata of a (P31 = instance of) "functional group", showing a regular SMILES with a "[*]" and a CXSMILES with the same but the "CX" addition "$_AP1$" causing the CDK-depiction to show a wigly line perpendicular to the bond "attaching the functional group" to the rest of the molecule.
@egonw@mastodon.social one things I learned about was the notion of "attachment points" in CXSMILES... works nicely in @wikidata :)
#CDK25UGM
16.03.2025 19:06 — 👍 1 🔁 1 💬 0 📌 0

cdk2024 #5: Chemistry Development Kit User Group Meeting - Day 2
Where the first workshop day had several talks about new and old features of the Chemistry Development Kit (CDK), the second day was a hackathon day. We hacked and we talked. The coding was not mostly only the CDK repository itself, but some things happened there too:
Some pointers:
* we worked on the Groovy Cheminformatics with the Chemistry Development Kit book
* move the repository to the GitHub organisation
* improved the build system
* it was explored how the CDK can generate SMILES for glycans
* continued work on updated tools using the CDK, e.g. ChemViz2
* code clean up, e.g. on JavaDoc and the XML parsing
* a JChemPaint release, based on CDK 2.10, with a flatpak for easy install on many Linux distributions
We further had discussions about a possible change of the license (what it would involve) and OSGi support. The problem there is that Java packages can only exist in one OSGi bundle, and this is currently not the discuss. We discussed that the current modules were partially setup to clean up dependencies, and generally modularize the CDK (e.g. each module could have a separate person responsible). We now want to propose a larger `core` module which covers the common cheminformatics functionality. A final discussion point I want to mention is that there are serious hints that we may have the Chemistry Development Kit as JavaScript in the browser soon!
I like to thank every one who joined the workshop, particularly those that travelled to Maastricht from the UK, Germany, and Bulgaria. Also thanks to the six participants online who joined on the first day!
the #CDK25UGM was great and exhausting. the rest of the week I did get around to writing up some notes from the second day, but I did now: https://doi.org/10.59350/n7e6r-p1e93
16.03.2025 18:02 — 👍 3 🔁 2 💬 1 📌 0
preview:
flatpak install --user https://dl.flathub.org/build-repo/169405/io.github.jchempaint.JChemPaint.flatpakref
11.03.2025 20:27 — 👍 0 🔁 0 💬 0 📌 0

Screenshot of JChemPaint with a SMILES with stereoinfo for the Pt for cis-platin and trans-platin
a beta release of JChemPaint 3.4 is now out: https://github.com/JChemPaint/jchempaint/releases/tag/3.4b
#CDK25UGM
11.03.2025 11:11 — 👍 1 🔁 2 💬 1 📌 0
after a great first @cdk user group meeting (see "cdk2024 #4: Chemistry Development Kit User Group Meeting - Day 1", https://doi.org/10.59350/e08pe-thb38), today we will have a hackathon (starting in about an hour)
#CDK25UGM
11.03.2025 06:53 — 👍 0 🔁 2 💬 1 📌 0
Chemistry Development Kit 2025 User Group Meeting
Repository to track the progress of the NWO Open Science Grant accepted for the CDK in 2023.
heading to Maastricht for a two day @cdk User Group Meeting #CDK25UGM
Rough schedule and list of speakers: https://cdk.github.io/nwo-openscience-2024/
If you like to listen in via Zoom, email me
#openscience #chemistry
10.03.2025 05:54 — 👍 4 🔁 6 💬 3 📌 0

Screenshot of a still of one of the videos, showing an editing session in the JChemPaint 2D chemical editor, with a highlighted alkane chain attached to a phenyl ring, about the be rotated.
the main JChemPaint branch has been rebased on CDK 2.10 and contains many new improvements. This PR contains a few short videos with some of the changes: https://github.com/JChemPaint/jchempaint/pull/199
#chemistry
04.03.2025 21:37 — 👍 1 🔁 3 💬 0 📌 0
Original post on fosstodon.org
new paper: "Extending Chemoinformatics Techniques With JMolecular Energy: A Robust CDK-Based Force Field Library" https://doi.org/10.1002/jcc.70071
" This paper introduces JMolecular Energy (JME), a novel, open-source Java library designed to implement MMFF94 with a robust and extendable API […]
28.02.2025 10:13 — 👍 2 🔁 6 💬 1 📌 0
Original post on fosstodon.org
On March 10-11 the @tgx_um group at Maastricht University (The Netherlands) is organizing a Chemistry Development Kit 2025 User Group Meeting.
Info and registration here: https://cdk.github.io/nwo-openscience-2024/
With six speakers on the first day, we have room for discussions on using the […]
24.02.2025 07:02 — 👍 1 🔁 1 💬 0 📌 0




![Screenshot of a record of Wikidata of a (P31 = instance of) "functional group", showing a regular SMILES with a "[*]" and a CXSMILES with the same but the "CX" addition "$_AP1$" causing the CDK-depiction to show a wigly line perpendicular to the bond "attaching the functional group" to the rest of the molecule.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:qj43mvyjyvzikd45py7f3q7s/bafkreib2h2h5lkocnyu2jtbdhvnksdy7djiry227lnr7uev7bjrelefpdm@jpeg)




