Big news! At least for me. I'll start building my own research group at the amazing IMBA in Vienna. Starting from single-cell omics data, we will develop computational tools and theoretical models to understand how single cells make decisions, in particular during development. I think that IMBA is
27.01.2026 13:44 —
👍 15
🔁 7
💬 2
📌 1
Stoked to share our latest work entitled: “Large-scale discovery of neural enhancers for cis-regulation therapies”
shorturl.at/H3Qww
This is an enormous team effort that I had the honour of spearheading with Nick Page and Florence Chardon.
Bluetorial below.
05.11.2025 15:09 —
👍 34
🔁 14
💬 2
📌 3
We are looking for a postdoc to study single-cell transcriptional heterogeneity in the human skin microbiome.
We have a new protocol mostly developed, but need someone to see it through. Experience with protocol dev or RNAseq appreciated.
Funded by a new grant from MIT-HEALS.
12.09.2025 16:49 —
👍 79
🔁 61
💬 0
📌 0


The are many anecdotes for #ChatGPT helping patients. But here's one with fallacious A.I. guidance that resulted in serious complications
www.acpjournals.org/doi/10.7326/...
11.08.2025 17:33 —
👍 82
🔁 30
💬 5
📌 1
Interested in #Bayesian #phylogenetics for analysing #single-cell lineage data? 🧬 To estimate timed trees 🕒, uncertainty and cell dynamic params!
I am teaching a BEAST 2 and TiDeTree tutorial during the #BC2 conference.
📍Basel, Sep 8, 2025
📅 Apply by Sep 1
Register here: bc2.ch/registration...
05.08.2025 22:55 —
👍 8
🔁 3
💬 0
📌 0
Interested in #Bayesian #phylogenetics for analysing #single-cell lineage data? 🧬 To estimate timed trees 🕒, uncertainty and cell dynamic params!
I am teaching a BEAST 2 and TiDeTree tutorial during the #BC2 conference.
📍Basel, Sep 8, 2025
📅 Apply by Sep 1
Register here: bc2.ch/registration...
05.08.2025 22:55 —
👍 8
🔁 3
💬 0
📌 0

Beeswarm plot of the prediction error across different methods of double perturbations showing that all methods (scGPT, scFoundation, UCE, scBERT, Geneformer, GEARS, and CPA) perform worse than the additive baseline.

Line plot of the true positive rate against the false discovery proportion showing that none of the methods is better at finding non additive interactions than simply predicting no change.
Our paper benchmarking foundation models for perturbation effect prediction is finally published 🎉🥳🎉
www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵
04.08.2025 13:52 —
👍 126
🔁 57
💬 2
📌 6
Our lab is helping with the organisation of this year's EMBO YSF. We have a great line up of invited speakers from (mostly) the wider Baltic region & a @nightsciencepod.bsky.social workshop on creativity in science (I did it a couple of years ago, it was awesome). Please share & consider joining us!
28.07.2025 15:48 —
👍 4
🔁 3
💬 0
📌 0
New Postdoc Fellowship Program Builds on Innovative SeattleHub Research | Brotman Baty Institute
BBI
POSTDOC ALERT: Applications now open for SeaBridge postdoc fellowship; opportunity to work on leading-edge biotech w/researchers at the Seattle Hub for Synthetic Biology.
brotmanbaty.org/news/new-pos...
@marionpepper.bsky.social
@jshendure.bsky.social
@coletrapnell.bsky.social
16.06.2025 16:36 —
👍 3
🔁 4
💬 0
📌 0
Application deadline extended for Taming the BEAST in Beijing!
Interested in #Bayesian #phylogenetics? 🧬
Join the Taming the BEAST workshop — theory + hands-on tutorials incl. #singleCell, #devBio & #lineageTracing!
📍Beijing, July 14–18, 2025
📅 Apply by April 25
I'll teach on TiDeTree & cell trees
🔗 taming-the-beast.org/news/Deadlin...
17.04.2025 19:28 —
👍 9
🔁 5
💬 0
📌 0
Application deadline extended for Taming the BEAST in Beijing!
Interested in #Bayesian #phylogenetics? 🧬
Join the Taming the BEAST workshop — theory + hands-on tutorials incl. #singleCell, #devBio & #lineageTracing!
📍Beijing, July 14–18, 2025
📅 Apply by April 25
I'll teach on TiDeTree & cell trees
🔗 taming-the-beast.org/news/Deadlin...
17.04.2025 19:28 —
👍 9
🔁 5
💬 0
📌 0

💡 Learn to analyze genetic lineage tracing data with TiDeTree in BEAST 2! Gain hands-on experience in setting up and running analyses, from preparing data to interpreting results.
🔹 Organized by @seidels.bsky.social
& Antoine Zwaans (@ethzurich.bsky.social)
👉 Register: bc2.ch/registration...
31.03.2025 13:06 —
👍 2
🔁 2
💬 1
📌 1
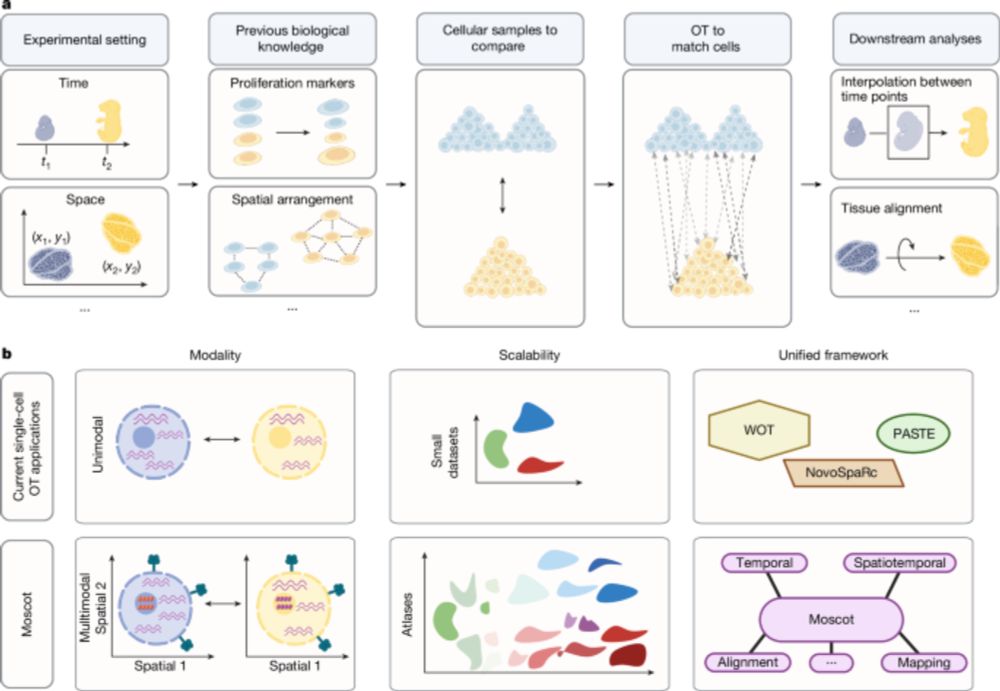
Mapping cells through time and space with moscot - Nature
Moscot is an optimal transport approach that overcomes current limitations of similar methods to enable multimodal, scalable and consistent single-cell analyses of datasets across spatial and temporal...
Good to see moscot-tools.org published in @nature.com ! We made existing Optimal Transport (OT) applications in single-cell genomics scalable and multimodal, added a novel spatiotemporal trajectory inference method and found exciting new biology in the pancreas! tinyurl.com/33zuwsep
23.01.2025 08:41 —
👍 49
🔁 13
💬 1
📌 3

We've wondered for a while why #gastruloids are so sensitive to initial cell numbers. Here www.biorxiv.org/content/10.1... led by U, Fiuza, we explore this & observe effects of a number of variables on final structure and think what this tells us @ embryogenesis
#IInNumbersWeTrust #NotInTheGenes
17.12.2024 18:52 —
👍 47
🔁 19
💬 1
📌 0
Call for papers - Spatial tumor heterogeneity
Weini Huang, @xiaofu90.bsky.social, Dmitrii Shek and I are excited to be guest editing a BMC Cancer collection on "Spatial tumor heterogeneity". We welcome studies ranging from spatial transcriptomics to math models. For details and submission guidelines see www.biomedcentral.com/collections/... 🧪
16.12.2024 19:08 —
👍 11
🔁 2
💬 1
📌 2

Spread the word! Registration for the "Mutations in Time and Space" conference is open. The meeting is all about the origins, patterns, and consequences of mutations across cells, individuals, populations, species. Abstract submission deadline is Jan 17th broadinstitute.swoogo.com/mits2025/597...
06.12.2024 14:29 —
👍 22
🔁 10
💬 1
📌 3
Would love to be added too!
04.12.2024 18:17 —
👍 0
🔁 0
💬 0
📌 0
Thank you for highlighting our review!
03.12.2024 13:05 —
👍 1
🔁 0
💬 0
📌 0

Interested in recording cell dynamics? Beautiful review from colleagues around Tanja Stadler and Martin Tran @NatureRevGenet on DNA recording in single cells and lienages. With lots of exciting opportunities for single cell modeling. nature.com/articles/s41576-024-00788-w
03.12.2024 09:30 —
👍 57
🔁 13
💬 1
📌 1
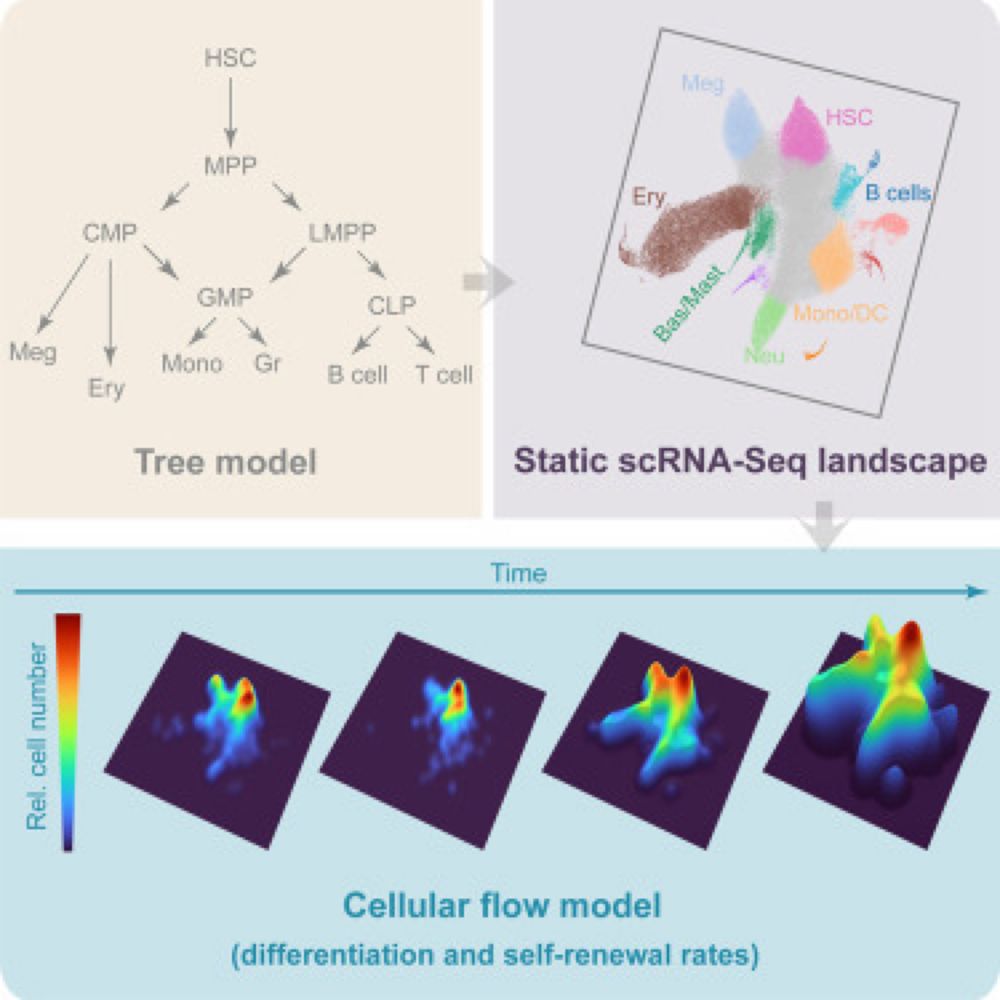
A time- and single-cell-resolved model of murine bone marrow hematopoiesis
The paradigmatic hematopoietic tree model is increasingly recognized to be limited, as it is based on heterogeneous populations largely defined by non…
Very happy to share our latest paper:”Time and single-cell resolved model of murine bone marrow hematopoiesis”:
www.sciencedirect.com/science/arti...
With the groups of Kamil Kranc and Donal O’Carroll, we have generated a new hematopoiesis model fit for the single cell genomics era.
05.01.2024 15:55 —
👍 10
🔁 1
💬 1
📌 0

‘DNA Typewriters’ Can Record a Cell’s History
Labs around the world are trying to turn cells into autobiographers, tracking their own development from embryos to adults.
Imagine a mouse in which all 10 billion cells keep a diary from fertilization. Imagine sentinel cells recording your experiences and sending out dispatches in the form of DNA. Here's a story I wrote about cell recorders 🧪 Gift link: nyti.ms/3Z3Mqza
25.11.2024 16:19 —
👍 109
🔁 33
💬 2
📌 9




















