Evaluating Outward Assembly on a Surprising Junction
Investigating a lab-contamination chimera that joins SARS-CoV-2 with a synthetic plasmid, this post shows how outward assembly quickly rebuilt ≈...
Overall, we find that outward assembly did really well: it recovered 959bp of the plasmid at 99.4% identity.
However, usability for time-sensitive investigations could be improved, which we’re planning to do this quarter.
Read more: naobservatory.org/blog/outward...
25.07.2025 16:32 — 👍 0 🔁 0 💬 0 📌 0
The suspicious sequence was a SARS-CoV-2 protein stuck to a synthetic construct.
We identified the construct as a manufactured plasmid with a known genome.
Since we knew the genome we were supposed to recover, we had a rare chance to evaluate outward assembly in the real world.
25.07.2025 16:32 — 👍 0 🔁 0 💬 1 📌 0
SecureBio’s NAO detection system flags suspicious reads, but doesn't recover the surrounding genome.
We can recover these genomes with our outward assembly pipeline.
We recently tested outward assembly on a flagged SARS-CoV-2/plasmid construct – it did extremely well!
25.07.2025 16:32 — 👍 0 🔁 0 💬 1 📌 0
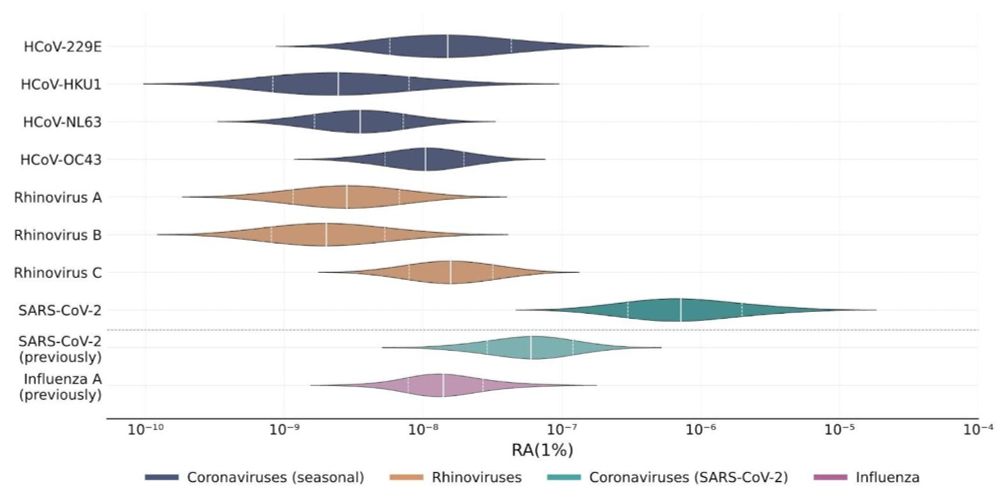
SARS-CoV-2 is highly detectable, but cold viruses are harder to detect.
We will refine these estimates as we generate more sequencing data.
naobservatory.org/blog/swab-ba...
10.07.2025 21:30 — 👍 0 🔁 0 💬 1 📌 0
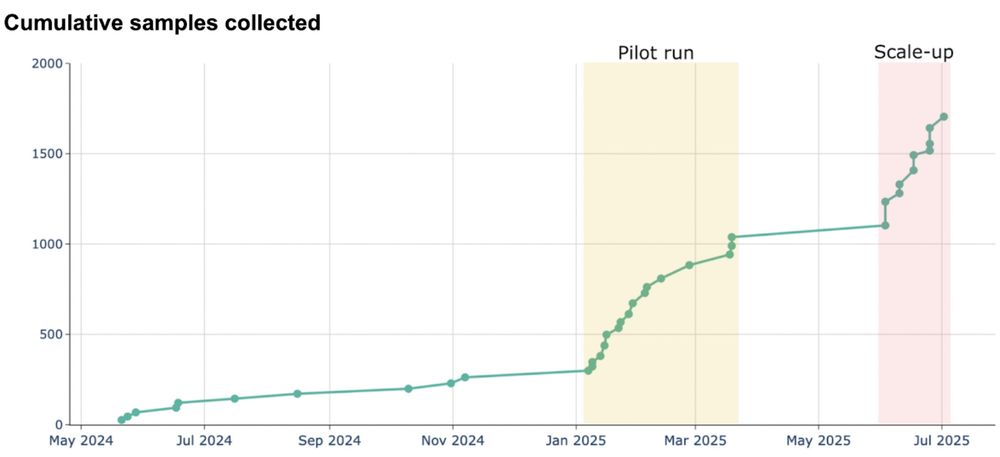
After our sampling sprint earlier this year, we’ve been scaling our Boston-based swab sampling program, hiring several field samplers.
This has allowed us to collect 100-200 swabs per day. Over the coming quarter we will scale to additional weekdays and further optimize sampling.
10.07.2025 21:30 — 👍 1 🔁 0 💬 1 📌 0
Our partners @solidevidence and Dave O'Connor have created new dashboards that present which pathogens we see in wastewater.
Find the dashboard here: dholab.github.io/public_viz/
10.07.2025 21:30 — 👍 0 🔁 0 💬 1 📌 0
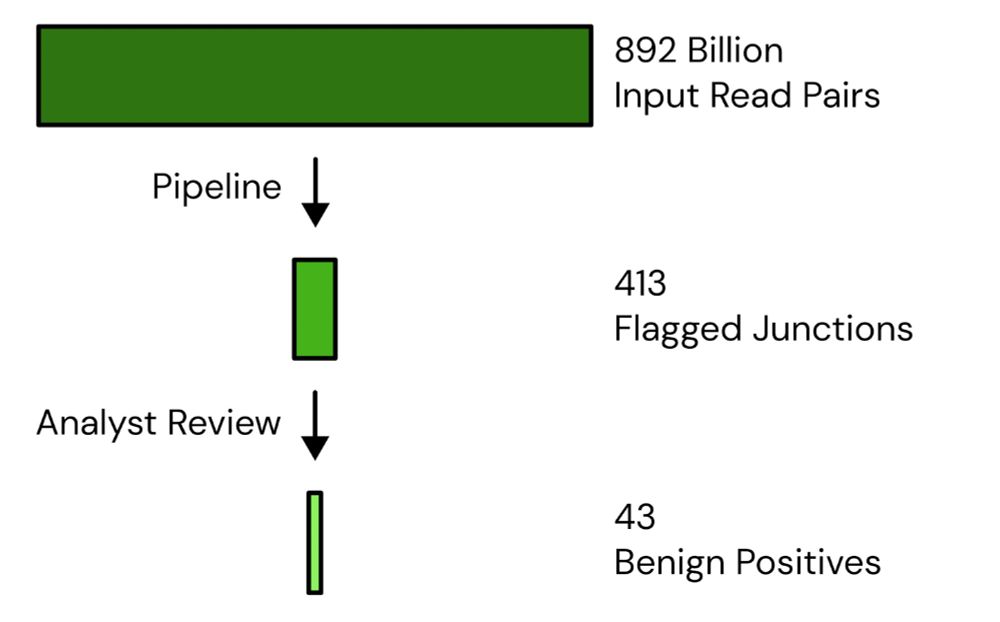
We’ve generated another 487B read pairs, through both our own sequencing, and through Marc Johnson’s lab (@solidevidence).
With the new data, our genetic engineering detection system has now analyzed 892B read pairs, flagging 413 chimeras, with 43 “benign positives”.
10.07.2025 21:30 — 👍 0 🔁 0 💬 1 📌 0

Nucleic Acid Observatory updates:
- We’ve further increased our sequencing capacity, producing 487B read pairs.
- Our partners created dashboards that summarize which pathogens we routinely see in wastewater.
- We’ve been scaling up our Boston-based swab sampling program.
10.07.2025 21:30 — 👍 3 🔁 0 💬 2 📌 0
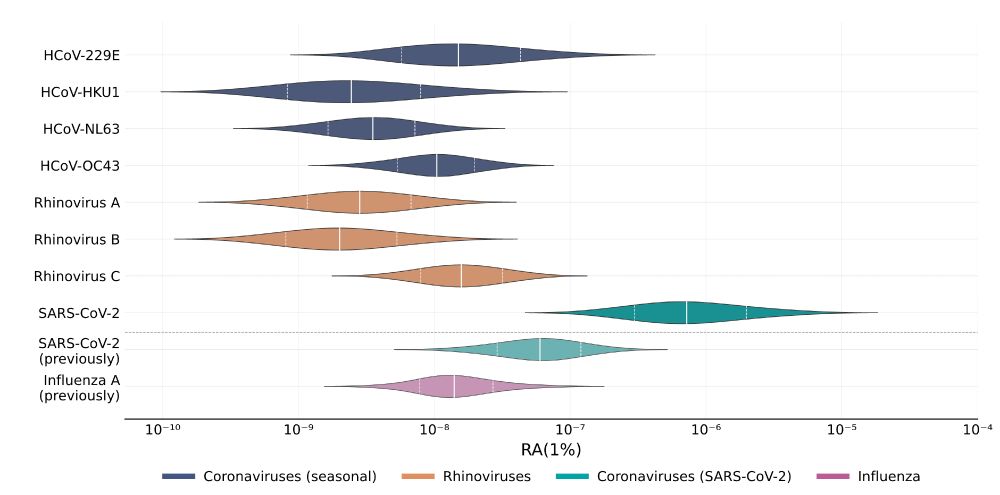
Linking this prevalence data with wastewater data, we estimated how easily different pathogens are detected in sequenced sewage.
SARS-CoV-2 was again confirmed to be readily detectable, but cold viruses showed relatively low detectability.
26.06.2025 13:50 — 👍 0 🔁 0 💬 1 📌 0
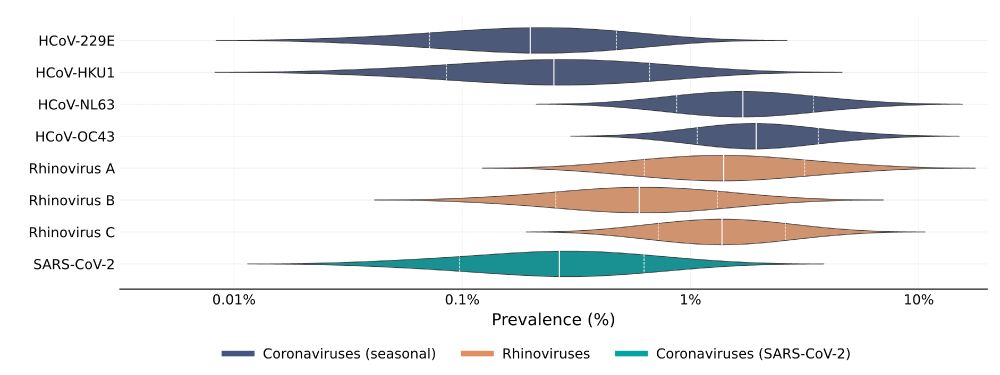
Using this we were able to estimate prevalence.
We found that cold viruses are highly prevalent in winter: Many have a prevalence of 1% or higher!
26.06.2025 13:50 — 👍 0 🔁 0 💬 1 📌 0
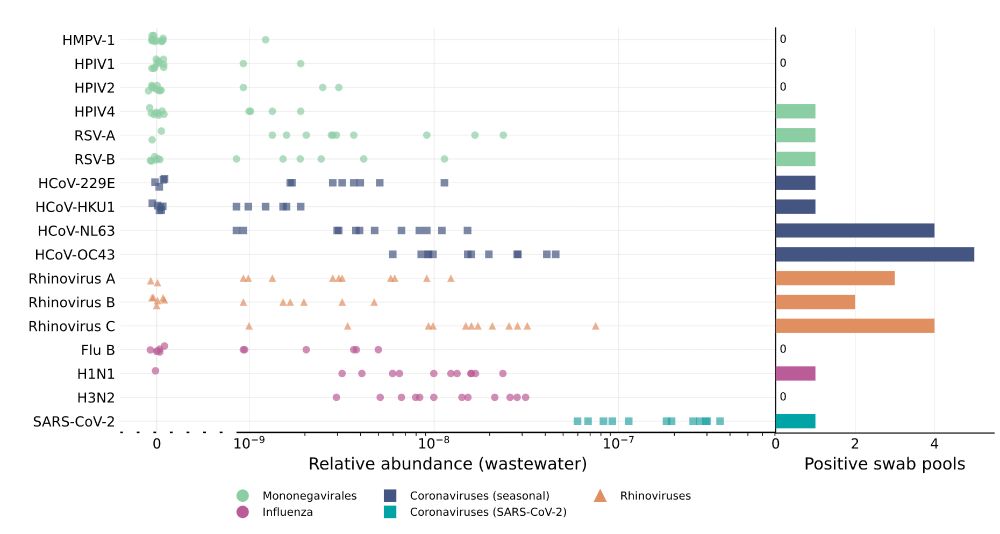
We created these estimates by analyzing nasal swabs, a recently added NAO sample stream.
Collecting, pooling, and sequencing nasal swabs, we got a lot of information about the presence of cold viruses in the population, which we then linked with paired wastewater data.
26.06.2025 13:50 — 👍 1 🔁 0 💬 1 📌 0

At the NAO, we’ve created new estimates on how well wastewater sequencing detects different pathogens.
SARS-CoV-2 is again highly detectable, but common cold viruses are harder to detect.
We will use this research to compare wastewater sequencing to other detection strategies.
26.06.2025 13:50 — 👍 10 🔁 4 💬 3 📌 0
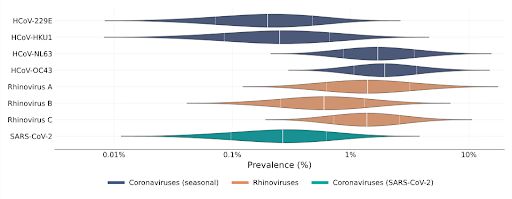
Chart of coronavirus and rhinovirus prevalence in the population
Using this we were able to estimate prevalence. We found that cold viruses are highly prevalent in winter: Many have a prevalence of 1% or higher!
19.06.2025 19:18 — 👍 0 🔁 0 💬 0 📌 0
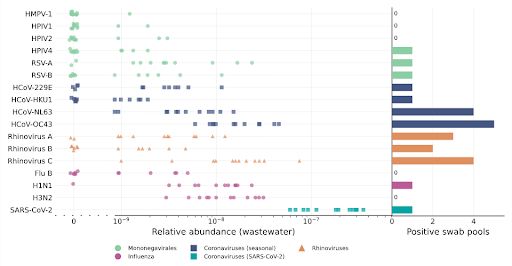
Chart of 1) relative abundance of various viruses in wastewater and 2) positive swab pools of those same viruses
We created these estimates by analyzing nasal swabs, a recently added NAO sample stream. Collecting, pooling, and sequencing nasal swabs, we got a lot of information about the presence of cold viruses in the population, which we then linked with paired wastewater data.
19.06.2025 19:18 — 👍 0 🔁 0 💬 1 📌 0
We’ll have two short talks, one by Dr. Mitchell Wolfe, former CDC Chief Medical Officer and now Ginkgo VP, and our own Jeff Kaufman, co-lead at the Nucleic Acid Observatory (NAO). Afterward, we’ll have ample time for 1-1 conversations.
18.06.2025 19:06 — 👍 0 🔁 0 💬 1 📌 0

Accelerating Biosecurity Networking Event · Luma
Join us for an exciting evening with Ginkgo Biosecurity and SecureBio as we explore how we can accelerate biosecurity efforts in the next 1-3 years. With…
There’s been a lot of action related to biosecurity recently (HHS potentially spending $52 million on a new “Biothreat Radar”, and both Ginkgo and the NAO scaling their surveillance efforts). If you want to discuss these developments, you can sign up here: lu.ma/igclw31d
18.06.2025 19:06 — 👍 0 🔁 0 💬 1 📌 0

SecureBio, partnering with Ginkgo Biosecurity will host an evening event on how to accelerate biosecurity on Thursday, July 31st, from 6:00-8:30 pm, in Boston (sign-up link in the next post).
18.06.2025 19:06 — 👍 1 🔁 0 💬 1 📌 0
Letter from the Executive Director
Some recent organizational updates
New Substack post from the Executive Director – an overview of some recent major accomplishments by our AI and biosurveillance teams, and some organizational updates.
substack.com/home/post/p-...
23.05.2025 15:38 — 👍 0 🔁 0 💬 0 📌 0

SecureBio | Substack
Click to read SecureBio, a Substack publication. Launched a month ago.
SecureBio recently launched a Substack! You can subscribe here: securebio.substack.com
Our Substack lets you get blog posts from the Nucleic Acid Observatory and AIxBio team directly to your inbox.
20.05.2025 18:50 — 👍 1 🔁 0 💬 0 📌 0
Pooled Lab Discards for Pathogen Detection
We’re excited about sampling strategies that can cheaply access many individual samples, approaching the coverage of municipal wastewater while ...
The medical system collects millions of samples for medical diagnoses. After testing, leftover samples could be repurposed for early pathogen detection.
@jefftk.com and @lennijusten.bsky.social explore sequencing pooled lab discards: naobservatory.org/blog/lab-dis...
16.05.2025 16:06 — 👍 0 🔁 0 💬 0 📌 0
Standard metagenomic assembly at our scale would be prohibitively expensive.
Instead, our approach searches billions of reads to find only those relevant to the seed, then assembles just those reads. 3/4
25.04.2025 17:28 — 👍 0 🔁 0 💬 1 📌 0
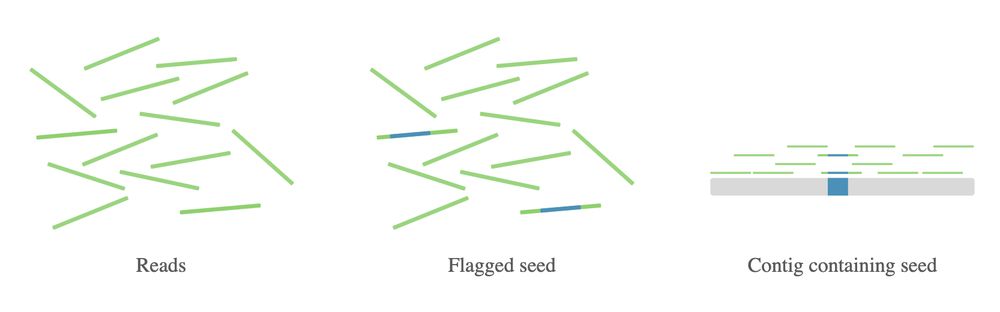
We've now developed outward assembly—a new open-source pipeline that efficiently builds longer sequences by extending outward from a suspicious "seed" sequence, like a read section flagged by our chimera detection pipeline. 2/4
25.04.2025 17:28 — 👍 0 🔁 0 💬 1 📌 0
At the Nucleic Acid Observatory, we do a *lot* of wastewater metagenomic sequencing—currently ~25B read pairs weekly, soon scaling to 75B.
Both finding suspicious sequences and understanding their genomic context are crucial challenges in our pathogen monitoring. 1/4
25.04.2025 17:28 — 👍 1 🔁 1 💬 1 📌 0
Thanks to the AI Safety Fund, which was extremely supportive and provided most of the funding.
And we want to highlight Jasper Götting, @jongsanders.bsky.social , and Pedro Medeiros from SecureBio who led the benchmark-building process. 13/13
23.04.2025 21:50 — 👍 0 🔁 0 💬 0 📌 0
[2] We suggest [expert practical assistance on tasks that could cause mass harm] should itself be considered a dual-use technology. Developers should consider "tiered access" for that capability, analogous to how we handle access to certain hazardous pathogens themselves. 12/13
23.04.2025 21:50 — 👍 0 🔁 0 💬 1 📌 0
So, what does this mean? Two quick take-homes.
[1] AIs can provide written troubleshooting better than virologists, according to standards set by the virologists themselves. 11/13
23.04.2025 21:50 — 👍 0 🔁 0 💬 1 📌 0

















