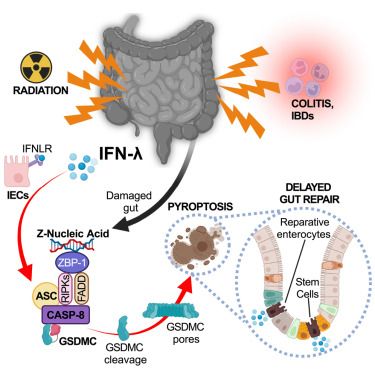
#InterferonPower! Thrilled for our latest work @cp-cell.bsky.social! With @danielboehmer.bsky.social, we dug into tons of papers & created what we hope will be a go-to resource for immunologists & non-immunologist about type I, II, III (& IV😉) #interferons! Free👉 authors.elsevier.com/a/1leKGL7PXu...
21.08.2025 15:20 —
👍 78
🔁 34
💬 1
📌 4

JSCBB Biotech Building in Boulder, CO
Are you bravely deciding to do a postdoc in the US? And also interested in some combination of genomics, immunology, and transposons? If so, consider applying to my lab at the BioFrontiers Institute in Boulder, Colorado!
jobs.colorado.edu/jobs/JobDeta...
🧪🧬 #TESky #interferosky
17.07.2025 19:36 —
👍 53
🔁 36
💬 1
📌 0
Pls. share widely
Calling all transposon fans & lovers of genetic innovation
MOBILE GENOME welcomes you in Heidelberg, Nov. 4–7 2025
→ Vibrant & friendly community
→ Cutting-edge talks from mechanisms to physiology
→ Plenty of surprises (TEs never stop innovating)
submit abstract by July 29
16.07.2025 08:43 —
👍 61
🔁 48
💬 1
📌 1
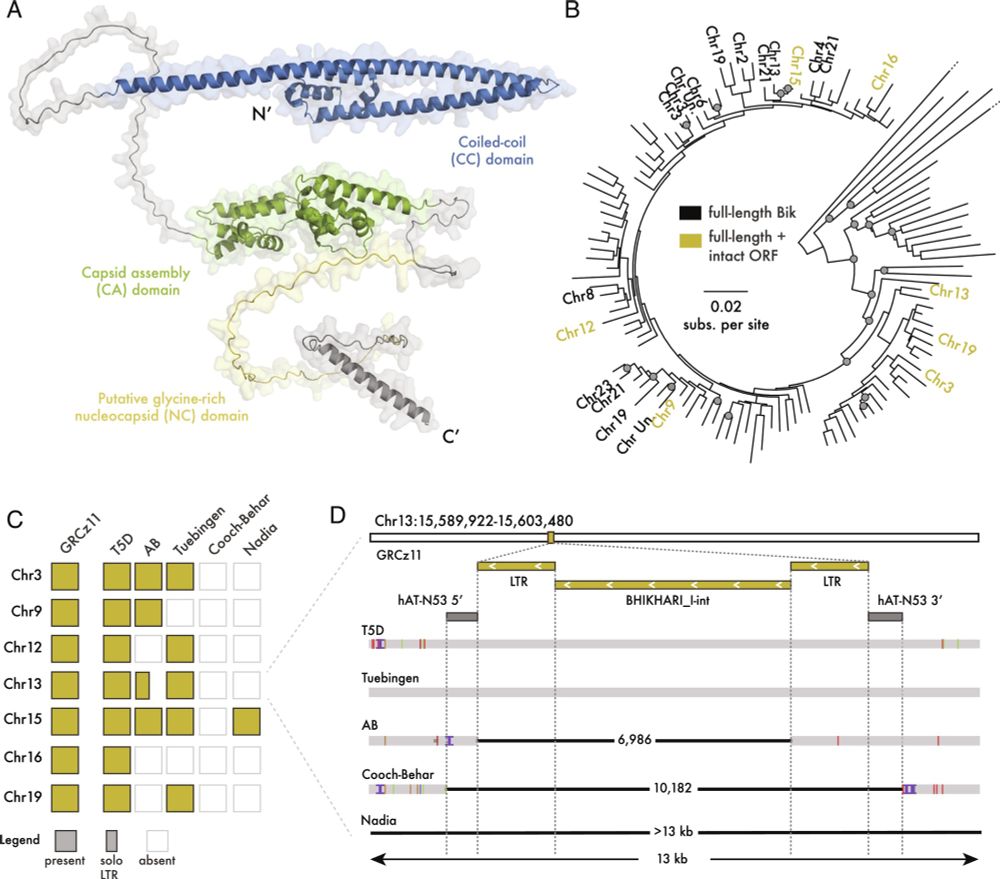
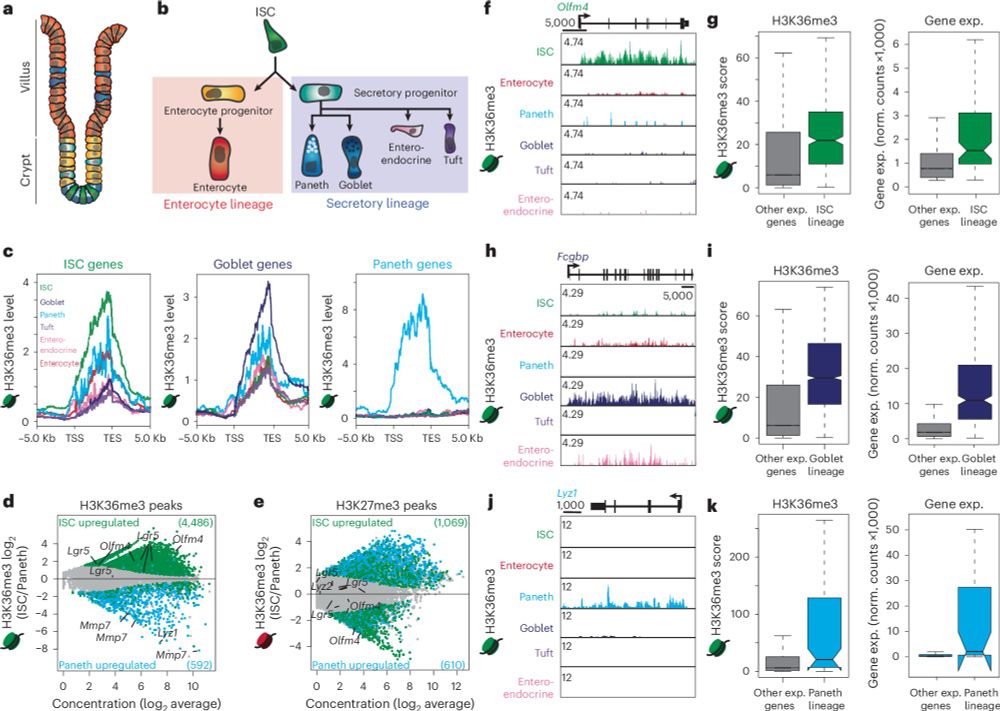
Gag proteins encoded by endogenous retroviruses are required for zebrafish development | PNAS
Transposable elements (TEs) make up the bulk of eukaryotic genomes and examples abound
of TE-derived sequences repurposed for organismal function. ...
💥🥳 At long last, our latest paper is out!
Gag proteins of endogenous retroviruses are required for zebrafish development
www.pnas.org/doi/10.1073/...
Led heroically by Sylvia Chang & @jonowells.bsky.social
A study which has changed the way I think of #transposons! No less! 🧵 1/n
30.04.2025 10:44 —
👍 259
🔁 108
💬 14
📌 21
If no single piece of the original ship remains in the current ship, is the ship still the Ship of Theseus?
And if one percent of all my body's cells get replaced every single day, is it really me who, two months ago, accepted your review request?
20.03.2025 06:00 —
👍 119
🔁 17
💬 6
📌 0
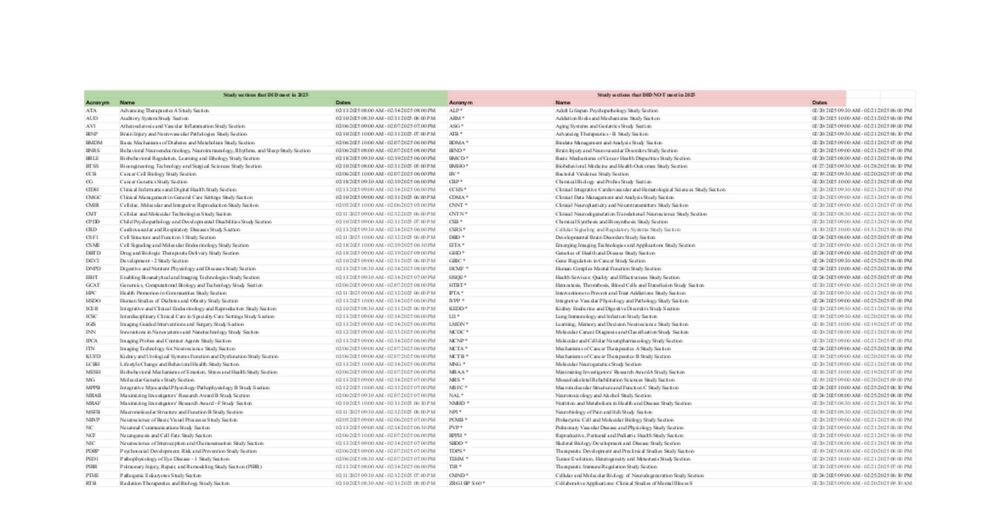
2025 Study section tracking
Wondering if your study section cancelled? I update this sheet daily. As of today, 56/124 study sections that should have met since Jan 2, 2025 have "not met as scheduled." docs.google.com/spreadsheets...
24.02.2025 16:17 —
👍 367
🔁 290
💬 44
📌 22
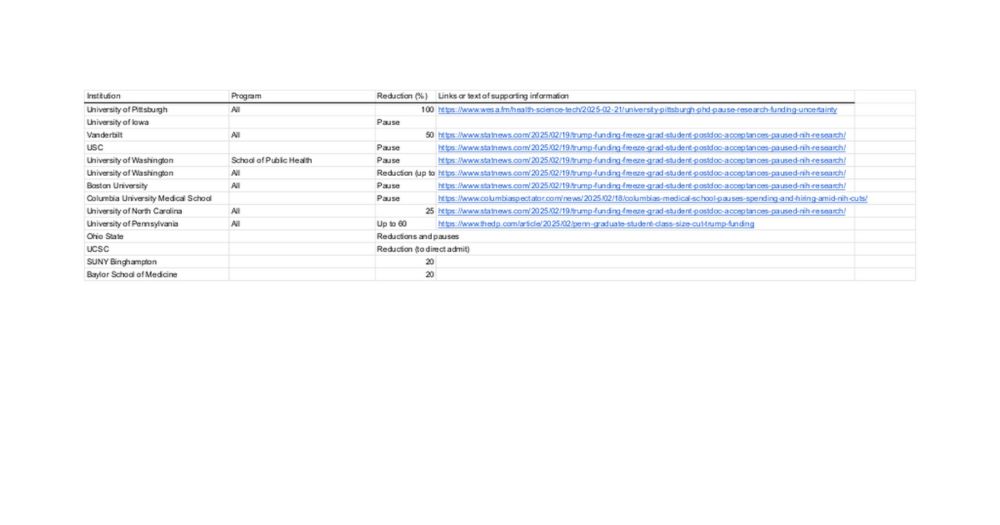
Graduate Reductions Across Biomedical Sciences (2025)
I created a brief spreadsheet of reductions I've heard of so far. Any additions you know of (especially if you have the links/receipts) would be great: docs.google.com/spreadsheets...
22.02.2025 13:23 —
👍 461
🔁 334
💬 38
📌 53
PREPRINT! Park et al. makes the case that we may be misunderstanding heterochromatin for past 30 years due to ChIP-Seq biases... 1/n
05.02.2025 18:34 —
👍 67
🔁 30
💬 5
📌 2
Congrats to the team, this is super important for studying TEs!
05.02.2025 19:43 —
👍 3
🔁 0
💬 1
📌 0
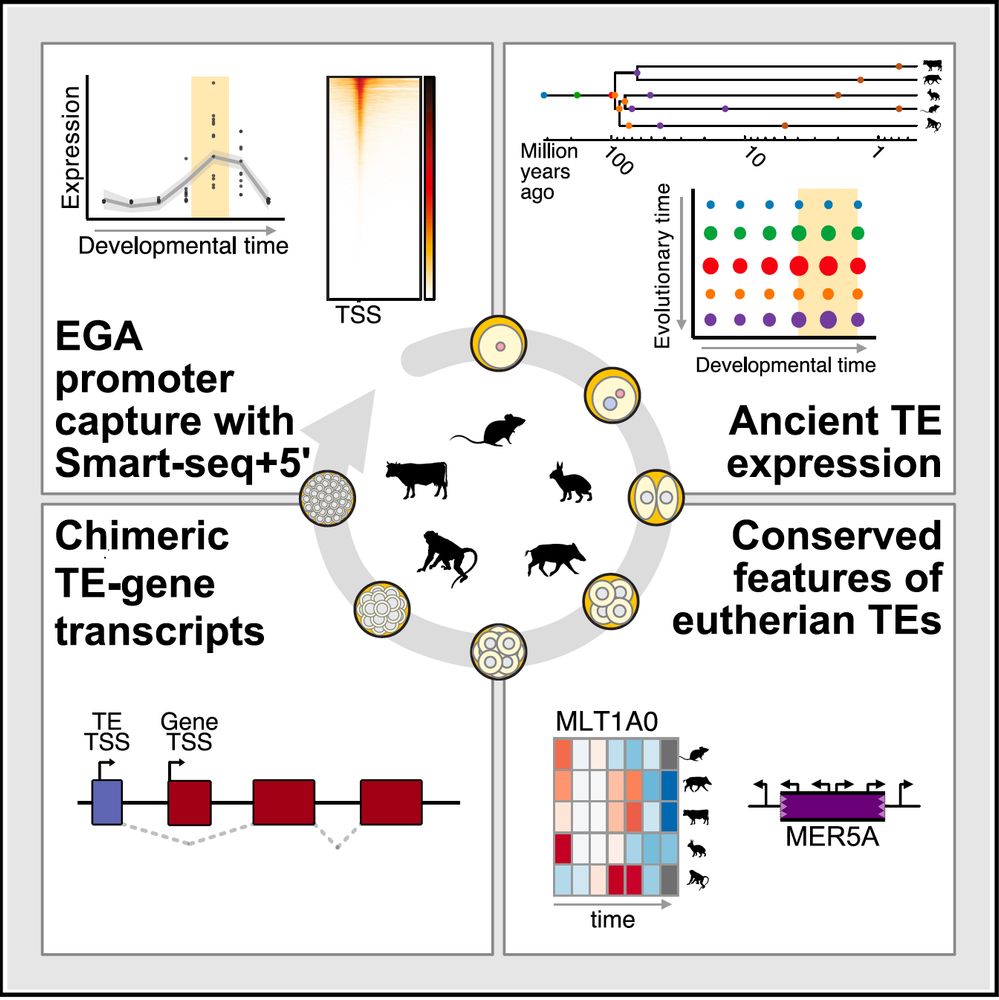
Very excited to share our paper on Gene and Transposable Element expression in mammalian preimplantation development, online today! www.cell.com/cell/fulltex...
A short thread to highlight some of our findings 🧵
20.01.2025 20:29 —
👍 114
🔁 52
💬 7
📌 5
Awesome, congrats to the team! 🎉
10.01.2025 16:37 —
👍 0
🔁 0
💬 1
📌 0
Postdoc: Molecular Biology/Genetics/Developmental Biology/Evolution/Computational Biology
I'm looking for a postdoc to join my fantastic team at the NIH in Bethesda, MD. Great resources. Competitive salary. www.training.nih.gov/jobs/pdf-mb-...
08.01.2025 19:06 —
👍 8
🔁 9
💬 0
📌 1
Check out this thread on the latest work from our lab, led by @emilykibby.bsky.social ! Here we used the power of computationally predicting protein-protein interactions to understand how a bacterial antiphage system senses infection.
19.12.2024 04:09 —
👍 43
🔁 15
💬 1
📌 0
First #bluetorial. 🎉🎉 We’re very happy to share our latest biorxiv #preprint, just in time before the holiday season. We explored how transposable elements (TEs) diversify eukaryotic proteomes and found a cool case in nematode F-box genes. #TEsky #evosky #Celegans
Short 🧵 with highlights.
19.12.2024 12:35 —
👍 74
🔁 27
💬 3
📌 7

Transposable element exonization generates a reservoir of evolving and functional protein isoforms
Transposable element exonization by unannotated splicing events produces stable protein isoforms with acquired functions that are subject to evolutionary selection.
#NotOnlyParasites! 2 Cell papers by @edchuong.bsky.social lab & Amigorena lab demonstrate that transposable elements #TEs can exonize giving rise to functional (often shorter) proteins with distinct functions: eg a decoy short IFNAR2 that inhibits #interferon signaling! www.cell.com/cell/fulltex...
12.12.2024 17:35 —
👍 26
🔁 16
💬 0
📌 0

All the credit for this discovery goes to postdoc @giuliapasquesi.bsky.social , who first noticed the isoform in her 2020 Lockdown Sideproject (tm). Her perseverance, creativity, and talent made this study possible and it was an honor to be a part of it. Watch out for her--she's on the job market! 😉
12.12.2024 18:56 —
👍 6
🔁 2
💬 1
📌 0

Beyond IFNAR2, our findings suggest that TE exonization may be a widespread yet hidden source of decoy isoforms that regulate immune signaling.
12.12.2024 18:56 —
👍 3
🔁 0
💬 1
📌 0

Our study shows how TE exonization gave rise to a primate-specific IFN decoy receptor, which acts as a dial to turn down IFN signaling in human cells. While IFN decoy receptors have evolved repeatedly in viruses (eg VACV B18), this is the first host-encoded IFN decoy receptor.
12.12.2024 18:56 —
👍 3
🔁 1
💬 2
📌 0

We also found that risk variants linked to severe COVID-19 in the IFNAR2 locus almost perfectly coincided with splicing QTLs associated with higher relative expression of the decoy. This suggests that variation in IFNAR2 splicing partly underlies individual variation in response to infection.
12.12.2024 18:56 —
👍 3
🔁 1
💬 1
📌 0

Giulia tested this by using CRISPR to dissect out each isoform in human cells, and did a battery of IFN assays. Whenever the short isoform was removed, cells showed more potent IFN responses, including immune gene activation, cytotoxicity, and antiviral effect against DENV-2 & SARS-CoV-2!
12.12.2024 18:56 —
👍 3
🔁 1
💬 1
📌 0

Giulia was surprised to see that the short isoform was ubiquitously expressed, often at >2x levels of the canonical isoform! Expression was conserved in other primates, and the protein was also detectable by mass-spec, suggesting that this isoform evolved an important immune function.
12.12.2024 18:56 —
👍 3
🔁 0
💬 1
📌 0


One caught her eye: a short isoform of the IFN receptor IFNAR2 formed by an exonized Alu. This isoform was first discovered over 20 yrs ago (Pfeffer et al 1997), yet has since been forgotten in the literature, presumably assumed to be silent in human cells.
12.12.2024 18:56 —
👍 3
🔁 1
💬 1
📌 0
Giulia revisited this assumption by digging into public long-read cDNA-seq, looking for exonized TEs that form isoforms with robust expression (>5 TPM) and mass-spec evidence. She uncovered hundreds of isoforms of protein-coding immune genes, which were mostly novel or poorly characterized.
12.12.2024 18:56 —
👍 2
🔁 0
💬 1
📌 0

Most introns are littered with TEs, which can become “exonized” and create cryptic isoform variants. Yet while thousands of exonization events have been detected, only a handful have been studied in detail, with most variants assumed to deleterious or nonfunctional.
12.12.2024 18:56 —
👍 4
🔁 0
💬 1
📌 0