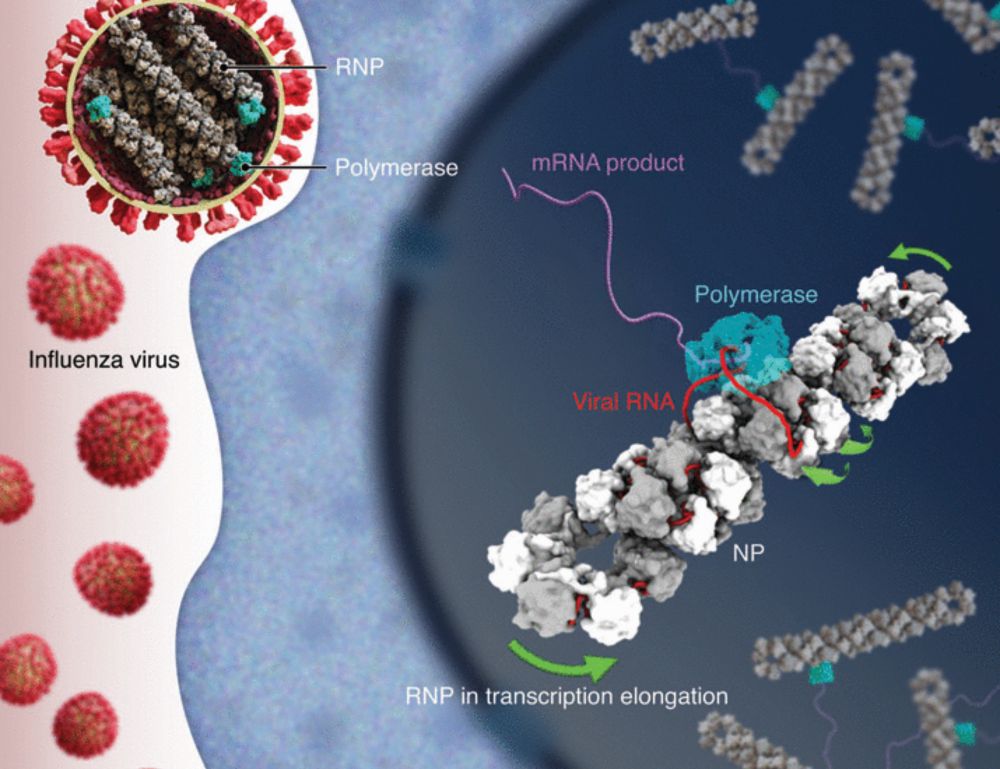
Anthony A. Hyman will become EMBL’s next Director General.
He joins EMBL from @mpi-cbg.de in Dresden. He is also Professor of Molecular Biology @tudresden.bsky.social, and was a group leader at EMBL Heidelberg from 1993 to 1999.
www.embl.org/news/people-...
27.11.2025 13:02 —
👍 253
🔁 67
💬 11
📌 11
Registration is open for "The complex life of #RNA" 2026 @embl.org in Heidelberg!
Excellent invited speakers and 30 short talks and even more flash talks selected from the abstracts!
20.11.2025 12:53 —
👍 31
🔁 18
💬 0
📌 0
Thanks a lot Elise!
07.11.2025 23:40 —
👍 1
🔁 0
💬 0
📌 0
The story of how multiple RBPs cooperate to repress translation is now out in @cp-cellreports.bsky.social by @marcopayr.bsky.social and in a great collaboration with @hennig-lab.bsky.social!
www.cell.com/cell-reports...
07.11.2025 15:34 —
👍 41
🔁 14
💬 3
📌 0

Excited to share my first PhD student’s @biorxivpreprint.bsky.social!
Tracking 5 dyes simultaneously Kavan Gor @embl.org tracks nascent #RNA folding during #ribosome assembly to correlate structural with functional information on single RNA molecules!
Check it out!
www.biorxiv.org/content/10.1...
01.11.2025 05:56 —
👍 85
🔁 25
💬 1
📌 3

Single-molecule multimodal timing of in vivo mRNA synthesis
mRNA synthesis requires extensive pre-mRNA maturation, the organisation of which remains unclear. Here, we directly sequence pre-mRNA without metabolic labelling or amplification to resolve transcript...
The timeline of co-transcriptional #mRNA processing and #modification may have to be revised: cleavage, termination, and #m6A deposition may occur earlier than most #splicing! Potential textbook changing study in bioRxiv by edueyras.bsky.social & Rippei Hayashi labs!
www.biorxiv.org/content/10.1...
29.04.2025 20:31 —
👍 55
🔁 21
💬 1
📌 1
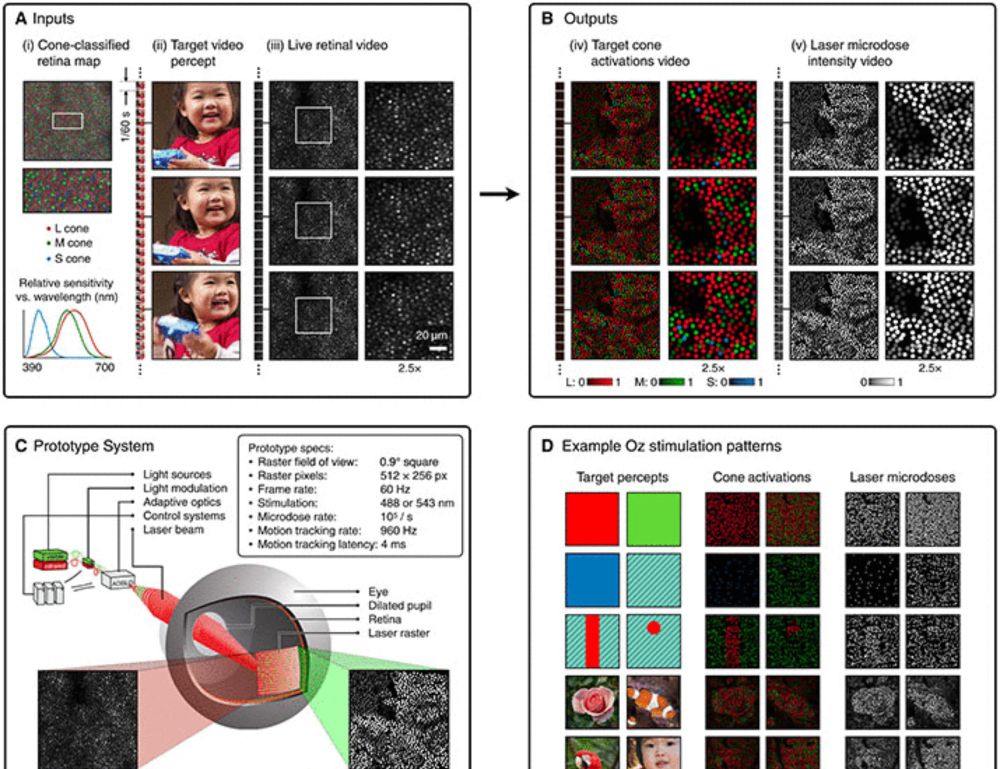
Novel color via stimulation of individual photoreceptors at population scale
Image display by cell-by-cell retina stimulation, enabling colors impossible to see under natural viewing.
Five people have seen a color never before visible to the naked human eye, thanks to a new retinal stimulation technique called Oz.
Learn more in #ScienceAdvances: scim.ag/442Hjn6
21.04.2025 17:56 —
👍 73
🔁 11
💬 4
📌 6
Thanks a lot for hosting me! Was very cool to talk to all of you!
17.04.2025 19:59 —
👍 1
🔁 0
💬 0
📌 0
Check out our preprint at BioRxiv revealing mechanisms of mRNP assembly during translation repression: (www.biorxiv.org/content/10.1...). Great collaboration with Olivier Duss (@olivierduss.bsky.social). Great Work Marco Payr (@marcopayr.bsky.social) and thanks for the video, explaining our paper.
11.04.2025 04:35 —
👍 36
🔁 14
💬 9
📌 1
Ever wondered how several individual RBPs cooperate to repress translation?
Lead by @marcopayr.bsky.social and in great collaboration with @hennig-lab.bsky.social, we simultaneously tracked the binding of several proteins to single mRNA molecules in real-time!
www.biorxiv.org/content/10.1...
12.04.2025 11:29 —
👍 53
🔁 17
💬 0
📌 3
Thanks Mark for the nice summary!
01.03.2025 21:26 —
👍 2
🔁 0
💬 0
📌 0

Title, authors and abstract from link
In a remarkable new paper, Qureshi and Duss use multi-colour single-molecule fluorescence microscopy to directly and simultaneously track transcription, translation & coupling 3/n
www.nature.com/articles/s41...
01.03.2025 13:40 —
👍 60
🔁 21
💬 3
📌 1
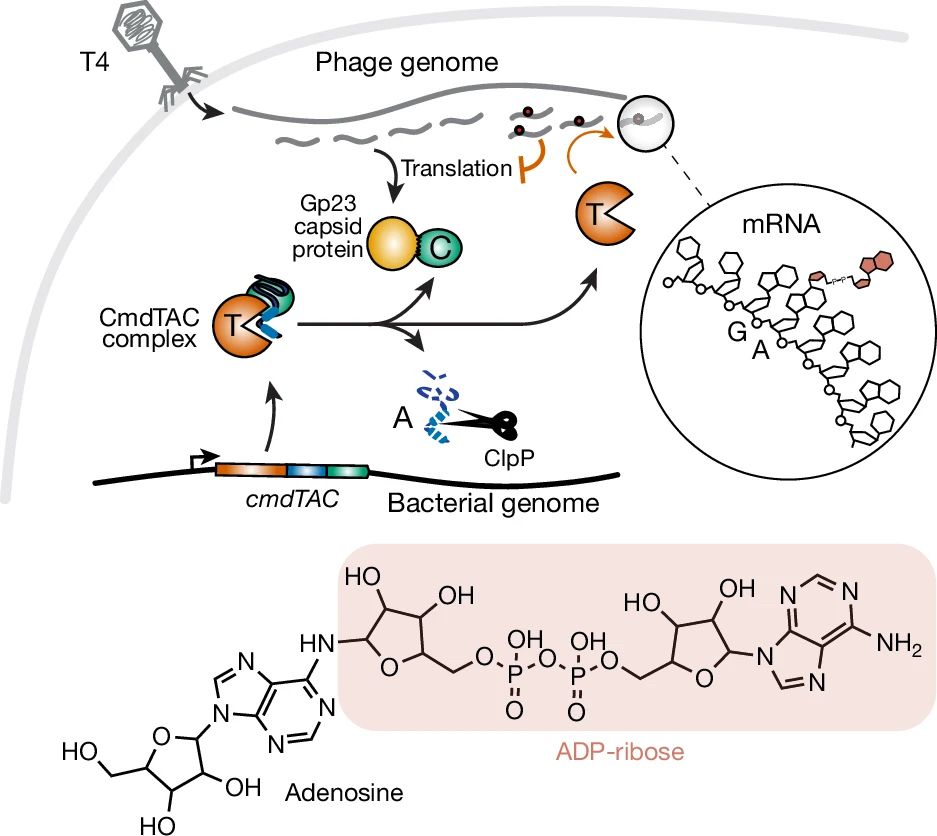
Graphic showing modification of mRNA with ADP-ribose mediated by bacterial enzyme cmdTAC
Check it out! Another novel RNA modification - ADP-ribosylation - that was previously only known on proteins. A cousin to #glycoRNA 👏
www.nature.com/articles/s41...
06.02.2025 19:38 —
👍 223
🔁 81
💬 3
📌 4
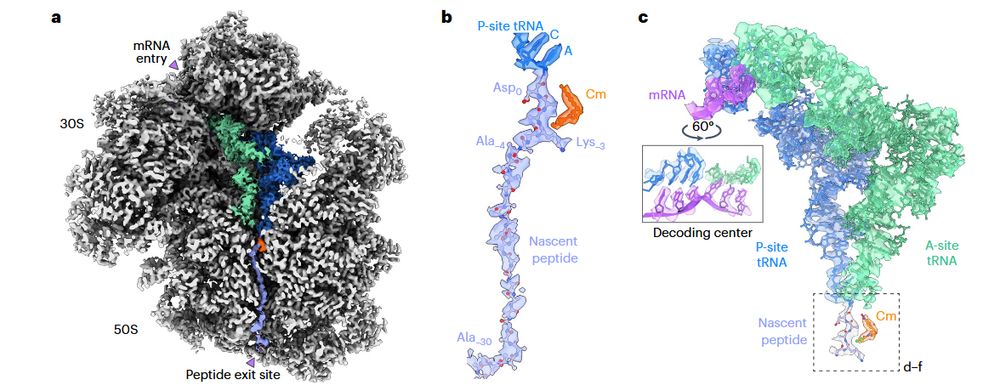
#cryoET of Mycoplasma pneumoniae Chloramphenicol-bound ribosomes at a global 3.0 Å resolution
www.nature.com/articles/s41...
12.12.2024 11:17 —
👍 37
🔁 13
💬 2
📌 1
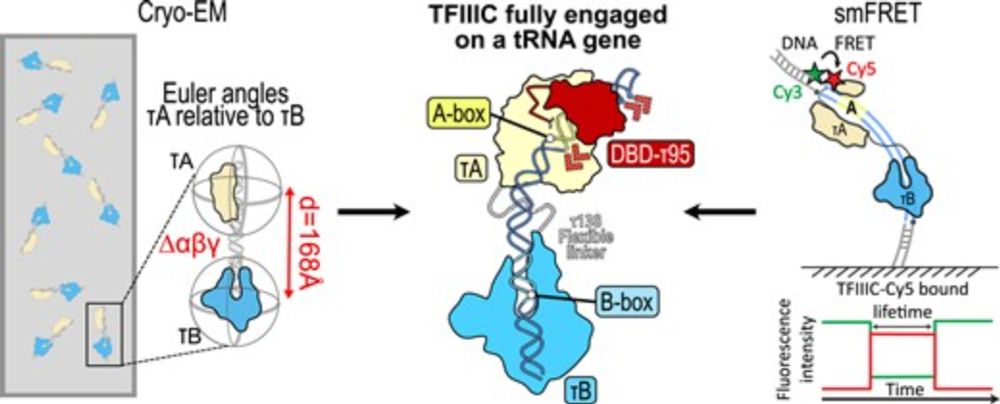
Structural and kinetic insights into tRNA promoter engagement by yeast general transcription factor TFIIIC
Abstract. Transcription of transfer RNA (tRNA) genes by RNA polymerase (Pol) III requires the general transcription factor IIIC (TFIIIC), which recognizes
Excited to share our new work on the earliest steps of Pol-III transcription: We provide structural and kinetic insights into tRNA promoter engagement by yeast transcription factor TFIIIC !
Cool collaboration with the Müller and Eustermann Labs @embl.org !
academic.oup.com/nar/advance-...
11.12.2024 07:39 —
👍 44
🔁 8
💬 1
📌 0
Thanks!
06.12.2024 18:15 —
👍 1
🔁 0
💬 0
📌 0
Thanks Arnaud!
06.12.2024 17:13 —
👍 0
🔁 0
💬 0
📌 0
Thanks Wolfgang!
06.12.2024 17:13 —
👍 0
🔁 0
💬 0
📌 0
Thanks!
06.12.2024 17:12 —
👍 0
🔁 0
💬 0
📌 0
Good to hear!
06.12.2024 17:12 —
👍 0
🔁 0
💬 0
📌 0
Thanks!
06.12.2024 17:11 —
👍 0
🔁 0
💬 0
📌 0
Thanks Tom!
06.12.2024 17:11 —
👍 0
🔁 0
💬 0
📌 0
Thanks David!
06.12.2024 17:11 —
👍 1
🔁 0
💬 0
📌 0