📢 Postdoc position: Cell Biology of Cyanobacteria
in my group, as part of the Excellence Cluster "Microbes for Climate" (M4C) in Marburg, Germany.
More information at shorturl.at/wNnDT (see Project 2)
🔗Apply at shorturl.at/VsEDl
📅Deadline: Nov 16, 2025
Please repost. #Postdoc
02.11.2025 19:13 — 👍 53 🔁 50 💬 2 📌 1

Learn how Bacillus siderophore benefit various plant species and the role of the biofilm matrix: our collaboration with Zhihui Xu's group published in @cp-cellreports.bsky.social
Siderophore-mediated iron enrichment in the biofilm matrix enhances plant iron nutrition
www.cell.com/cell-reports...
28.10.2025 07:52 — 👍 16 🔁 2 💬 1 📌 0
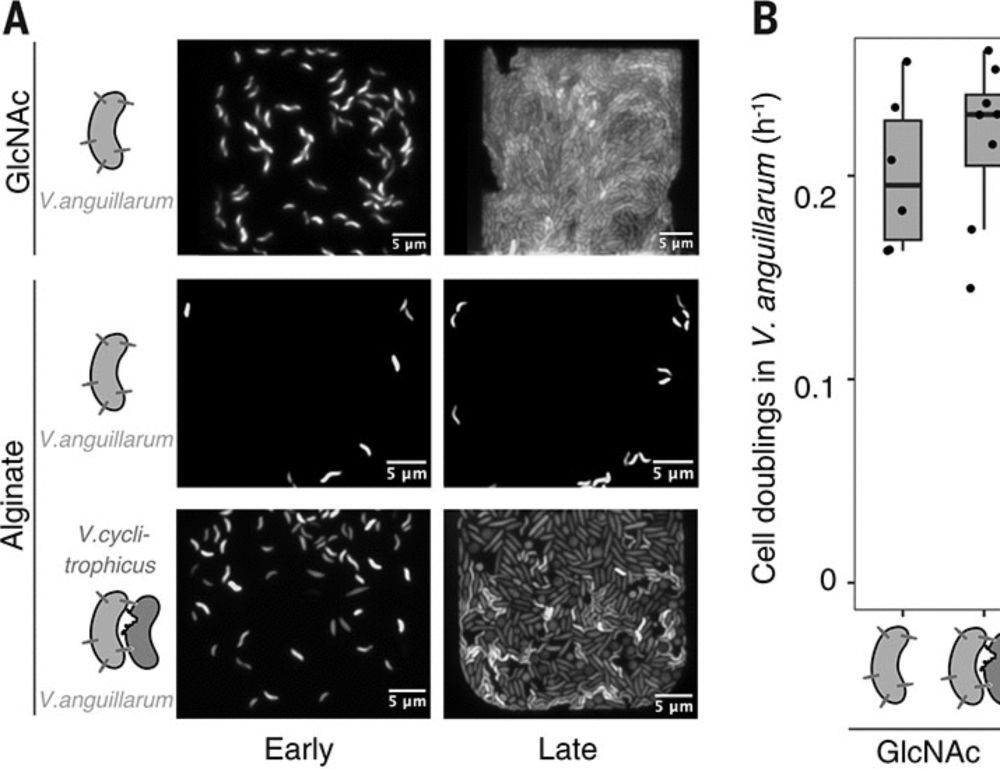
Who would have thought: nitrogen-fixing vibrios 😱 that interact with Pokkali rice roots and promote plant growth in brackish water. Amazing! Wonderful to see this detailed and careful investigation out for all to oogle. Many congratulations Ramesh 🎉
journals.asm.org/doi/10.1128/...
28.10.2025 08:08 — 👍 3 🔁 1 💬 0 📌 0

Genome-wide screen to identify Escherichia coli gene knockouts hypersensitive to trimethoprim and chloramphenicol. Top left: Schematic showing the screening strategy for identifying hypersensitive gene knockouts from the Keio mutant library. All mutants were cultured in LB and LB supplemented with trimethoprim (TMP) and chloramphenicol (CMP) at their respective IC50s. Top right: Distribution of drug-susceptibilities of Keio mutants grown in indicated growth media obtained after normalizing the growth of each mutant to wild type (set to 1). Mean ± S.D. derived after fitting each distribution to a Gaussian function are shown. Bottom left: Analysis of gene functional categories for hypersensitive mutants compared with the frequency of genes in each category encoded by E. coli K-12 MG1655. Percentage of the total in each case is provided above each bar. Bottom right: Comparison of hypersensitive mutants identified in the trimethoprim and chloramphenicol screens shown as a Venn diagram. Representative genes from each set are named and gene names are colored by functional categories.
Intrinsic resistance pathways & #ResistanceBreaking. Study shows that inhibiting efflux pumps & cell envelope biogenesis sensitizes #bacteria to #antibiotics, but rapid evolutionary recovery may limit long-term effectiveness of resistance-breaking strategies #AMR @plosbiology.org 🧪 plos.io/4huAtvo
23.10.2025 12:58 — 👍 11 🔁 7 💬 2 📌 1
Multiple small, curved, predatory bacteria (Bdellovibrio bacteriovorus) lyse the bacterial prey remnants below. How are they doing this & which predator proteins are involved in this process?
See our newly released research article 📑
=> rdcu.be/eL6gu
🦠 #microsky #bacteria #PredatoryBacteria
23.10.2025 16:44 — 👍 22 🔁 9 💬 2 📌 0

#microsky #mevosky @spp2389.bsky.social
A PhD position is available in my lab to work on:
Emergence and self-organisation of bacterial metabolism in consortia of cross-feeding bacteria.
Please RT
Deadline: 12.11.25
More infos 👇
shorturl.at/rAKAT
15.10.2025 13:07 — 👍 41 🔁 52 💬 1 📌 1
Big thanks to Rekha (lead), an absolute rockstar who drove the project, and Jyotsna for pushing this work forward!
Also grateful to our collaborator Prof. Siva Umapathy for the Raman spectroscopy expertise.
03.09.2025 20:03 — 👍 0 🔁 0 💬 0 📌 0
This is one of the first demonstrations linking biochemical markers of spores to their germination efficiency, using a non-invasive, label-free approach.
03.09.2025 20:03 — 👍 0 🔁 0 💬 1 📌 0
Most excitingly, these Raman spectral features of mature spores correlated strongly with germination efficiency across natural soil isolates of M. xanthus.
Meaning: Raman spectra can serve as predictive markers of germination success.
03.09.2025 20:03 — 👍 1 🔁 0 💬 1 📌 0
We found that:
1. Lipid and protein signatures shift dynamically during sporulation
2. Carotenoids accumulate in spores (stress protection!)
3. Nucleic acid signatures become compacted
03.09.2025 20:03 — 👍 1 🔁 0 💬 1 📌 0
Using single-cell Raman spectroscopy, we tracked key biochemical changes (lipids, proteins, nucleic acids, carotenoids) as M. xanthus transitioned from vegetative cells into spores.
03.09.2025 20:03 — 👍 0 🔁 0 💬 1 📌 0
Dormancy is one of the most widespread microbial survival strategies — from spores to persisters to VBNCs. But how do we predict which spores will successfully germinate and “wake up” again?
03.09.2025 20:03 — 👍 0 🔁 0 💬 1 📌 0

Raman Spectral Predictors of Spore Germination Efficiency in Natural Isolates of Myxococcus xanthus
Due to the prevalence and importance of dormant microbial forms in regulating microbial ecosystems, the generation of dormant structures, like spores, has been extensively studied. However, several aspects of the exit of bacterial spores from dormancy, i.e., the germination of spores, remain relatively unclear. Since the biology of cells before transitioning into spores and the biology of the spores themselves could potentially influence the germination process, we used Myxococcus xanthus as a model organism to demonstrate that Raman spectroscopy can be used to characterize the factors that affect the ability of individual cells and spores to sporulate and germinate, respectively. M. xanthus is a Gram-negative soil bacterium that forms spore-filled multicellular fruiting bodies upon starvation. Single-cell Raman spectral profiling revealed lower Raman peak intensities of nucleic acids as a marker for the beginning of sporulation. Moreover, Raman profiles of sporulating cells of M. xanthus demonstrated that the lipid peaks increased during the initial sporulation phase before decreasing during the late sporulation phase. We also observed higher carotenoid peaks in spores than in cells, which might explain the reason for spores being more tolerant to oxidative stress than the cells. Significantly, the trends in Raman bands of nucleic acids and proteins observed in the lab strain were also observed in the natural isolates. Furthermore, partial least squares regression (PLSR) analysis of peak intensities of the most significantly affected chemical groups demonstrated a strong correlation between Raman spectra and germination efficiencies of spores, suggesting that such spectral markers are potential indicators of the germination efficiency of the spore population.
New paper out in @pubs.acs.org Analytical Chemistry!
We show that Raman spectroscopy can identify biochemical signatures of spores in Myxococcus xanthus, and that these signatures can predict germination efficiency across natural isolates.
pubs.acs.org/doi/10.1021/...
03.09.2025 20:03 — 👍 1 🔁 1 💬 2 📌 0
Not many positions offer this rare combination: exciting research questions, a highly supportive work atmosphere, and an exceptional mentor
@KostChristian
. If you’re in theoretical biology and want to collaborate closely with experimentalists, this is it!
11.08.2025 04:50 — 👍 5 🔁 3 💬 1 📌 0
Congrats Glen. This sounds interesting. Look forward to reading it.
13.06.2025 12:10 — 👍 0 🔁 0 💬 1 📌 0
Big kudos to @sahelis.bsky.social , Sneha, and @vaibhav-sharma.bsky.social ! Stay tuned for more stories on how microbial predation affects the evolution of resistance in pristine environments.
04.06.2025 16:13 — 👍 0 🔁 0 💬 0 📌 0
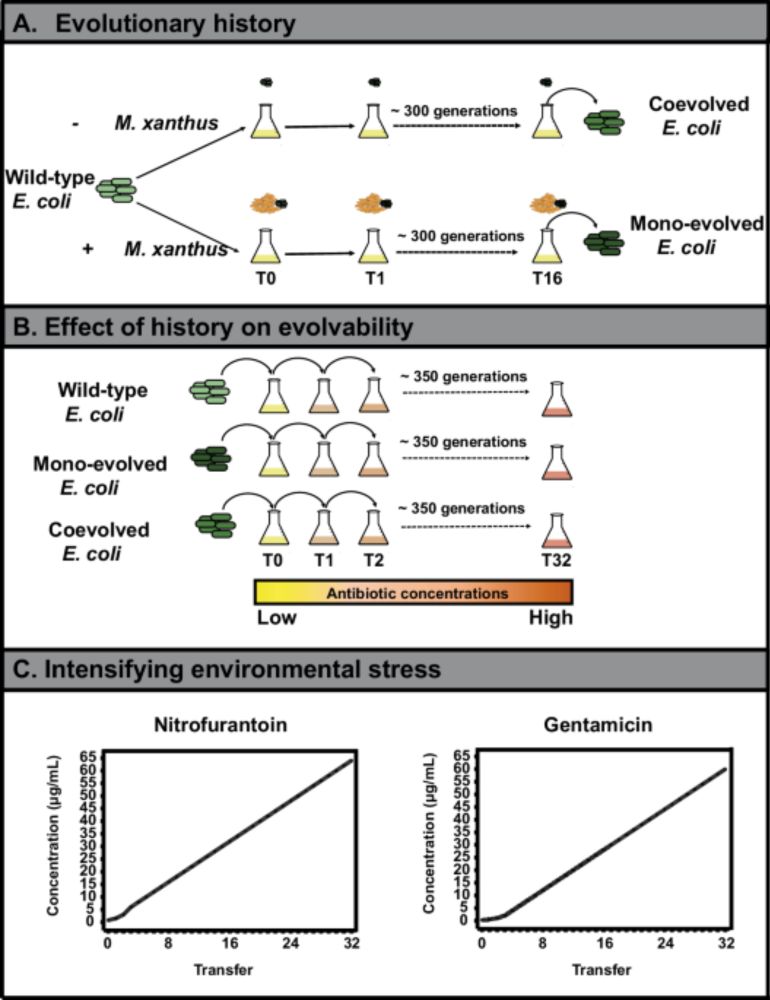
Thus, microbial interactions in pristine environments can influence the evolvability of pathogens long before clinical antibiotic exposure.
04.06.2025 16:13 — 👍 0 🔁 0 💬 1 📌 0
These findings are important because: Resistance traits arising under biotic stress may be less constrained, and potentially more evolutionarily potent, than those shaped in antibiotic-only settings.
04.06.2025 16:13 — 👍 0 🔁 0 💬 1 📌 0
Our work demonstrates that coevolutionary history with a predator influences not only present-day survival but also the trajectory, speed, and cost of future adaptations, such as antibiotic resistance.
04.06.2025 16:13 — 👍 0 🔁 0 💬 1 📌 0
While abiotic influences on evolvability have been widely studied, the historical contingencies of antagonisms, such as microbial predation, remain underexplored.
04.06.2025 16:13 — 👍 0 🔁 0 💬 1 📌 0
Anti-predatory adaptations in E. coli were retained even after propagation in predator-free environments. These historical imprints persist and affect future evolutionary responses.
04.06.2025 16:13 — 👍 0 🔁 0 💬 1 📌 0
And yet, if resistance does evolve in predator-adapted prey, it comes at a lower fitness cost than in other backgrounds. This means such resistance may spread faster in microbial populations.
04.06.2025 16:13 — 👍 0 🔁 0 💬 1 📌 0
Key finding: Predation history restricts the evolution of high-level antibiotic resistance in prey bacteria, compared to those evolved only under abiotic conditions or wild-type controls.
04.06.2025 16:13 — 👍 0 🔁 0 💬 1 📌 0
We show that E. coli strains previously exposed to M. xanthus showed altered survivability when challenged with increasing concentrations of antibiotics.
04.06.2025 16:13 — 👍 0 🔁 0 💬 1 📌 0
Early career PI and mother
Microbiologist and geneticist.
Evolutionary systems/cell biologist. EMBO and SNSF Postdoctoral Fellow with Dmitri Petrov and Dan Jarosz at Stanford. PhD with Andreas Wagner at the University of Zurich. Studying how molecular and cellular systems shape, and are shaped by, evolution.
Figuring out how microbes get along (or don’t).
Ph.D. student in Kost lab, University of Osnabrück.
Doctoral Researcher @Kost lab || Universitat Osnabrück || IIT KGP-IACS Kolkata
Senior Scientist, Microbiologist
Interdisciplinary scientist interested in microbial communities.
Postdoc at Max Planck Institute for Evolutionary Biology
https://kaumudiprabhakara.github.io/
Formerly: postdoc at University of Chicago
PhD: Cornell University
IZTECH-Chemistry, I like to be Polymath(learning different subjects), Polygot(interested in the learning different languages) comments and opinions are my own RT≠ is not endorsement (he/him)
Biologist at Dalhousie University in Nova Scotia. Researcher of genomes and evolution. Salido de la Universidad de Buenos Aires. 🇦🇷🇨🇦
Professor, Biology, Indiana University
Bacterial physiology and cross-feeding interactions
Artist: https://bit.ly/3oy86jf
Sheltering under solar panels.
Posts are my own.
Multicellular and symbiotic evolutionary biologist. Postdoc at University of Oxford. PhD from WUSTL
scientist 👩🏻🔬 | planetary health @ ulisboa | post = mine ≠ endorse
Interested in microbial ecology & evolution. Views are only my own. (he/him)
🎓: https://scholar.google.com/citations?hl=en&user=OBPpZq4AAAAJ&view_op=list_works&sortby=pubdate
👨💻: https://github.com/raufs
Professor at UArizona interested in genome rummaging, microbial evolution, and occasional tales about having backyard farm animals.
“I’m something of a scientist myself”
Baltrus(at)Arizona.edu
PhD student
Bacterial Ecology and Evolution Lab
Indian Institute of Science
A Simons collaboration which investigates the principles underpinning the self-organization, structure, and function of #microbial communities in the #ocean.
Asst. Professor, at Indian institute of Science, Bangalore. Biochemist and molecular biologist interested in #smallRNAs and host #parasite interactions. #newPI
A student of science -trying many things at a time
Postdoc @UPenn
Postdoc & PhD @UniZurich
MSc & BSc @PresidencyKolkata