Klebsiella pneumoniae genomics tutorials – KlebNET-GSP
The @klebnet.bsky.social team are pleased to share slides from our “Klebsiella pneumoniae Genomic Epidemiology & Antimicrobial Resistance” lecture series!
Topics include Kleb diversity, lineages, AMR, hypervirulence, how to use Kaptive & Kleborate for typing, and more!
klebnet.org/2025/11/18/k...
18.11.2025 08:25 — 👍 33 🔁 14 💬 1 📌 1
Thrilled to share this preprint on Klebsiella pneumoniae plasmids in a One Health setting 🦠 Fantastic work by @twinkler.bsky.social! 🎉 @irenlohr.bsky.social @margaretlam.bsky.social
16.09.2025 09:40 — 👍 19 🔁 11 💬 0 📌 0
Annual Norwegian NORM/NORM-VET report released.
Norway’s strict antibiotic policies pay off. Record-low use in humans & animals, and some of Europe’s lowest #AMR rates. But ESBL & VRE are on the rise, so vigilance is vital.
www.unn.no/4a675d/sitea...
16.09.2025 17:34 — 👍 34 🔁 12 💬 0 📌 1
Thrilled to share this preprint on Klebsiella pneumoniae plasmids in a One Health setting 🦠 Fantastic work by @twinkler.bsky.social! 🎉 @irenlohr.bsky.social @margaretlam.bsky.social
16.09.2025 09:40 — 👍 19 🔁 11 💬 0 📌 0

Excited to share the first beta release of AMRrules at #ABPHM! (Poster 42 tonight)
interpretamr.github.io/AMRrules
22.05.2025 09:35 — 👍 42 🔁 32 💬 5 📌 4
Delighted to see this paper from danderson123.bsky.social 's PhD out. We have been building tools for AMR gene detection for over a decade now, but multicopy genes remain challenging. Dan shows that with a gene-space de Bruijn graph and long reads, you can do well
www.biorxiv.org/content/10.1...
19.05.2025 09:28 — 👍 89 🔁 50 💬 4 📌 4

Fast and Accurate in silico Antigen Typing with Kaptive 3
Surface polysaccharides are common antigens in priority pathogens and therefore attractive targets for novel control strategies such as vaccines, monoclonal antibody and phage therapies. Distinct serotypes correspond to diverse polysaccharide structures that are encoded by distinct biosynthesis gene clusters, e.g. the Klebsiella pneumoniae species complex (KpSC) K- and O- loci encode the synthesis machinery for the capsule (K) and outer-lipopolysaccharides (O), respectively. We previously presented Kaptive and Kaptive 2, programs to identify K and O-loci directly from KpSC genome assemblies (later adapted for Acinetobacter baumannii), enabling sero-epidemiological analyses to guide vaccine and phage therapy development. However, for some KpSC genome collections, Kaptive (v≤2) was unable to type a high proportion of K-loci. Here we identify the cause of this issue as assembly fragmentation, and present a new version of Kaptive (v3) to circumvent this problem, reduce processing times and simplify output interpretation. We compared the performance of Kaptive v2 and Kaptive v3 for typing genome assemblies generated from subsampled Illumina read sets (decrements of 10x depth), for which a corresponding high quality completed genome was also available to determine the 'true' loci (n=549 KpSC, n=198 A. baumannii). Both versions of Kaptive showed high rates of agreement to the matched true locus among 'typeable' locus calls (≥96% for ≥20x read depth), but Kaptive v3 was more sensitive, particularly for low depth assemblies (at <40x depth, v3 ranged 0.85-1 vs v2 0.09-0.94) and/or typing KpSC K-loci (e.g. 0.97 vs 0.82 for non-subsampled assemblies). Overall, Kaptive v3 was also associated with a higher rate of optimal outcomes i.e. loci matching those in the reference database were correctly typed and genuine novel loci were reported as untypeable (73-98% for v3 vs 7-77% for v2 for KpSC K-loci). Kaptive v3 was >1 order of magnitude faster than Kaptive v2 making it easy to analyse thousands of assemblies on a desktop computer, facilitating broadly accessible in silico serotyping that is both accurate and sensitive. The Kaptive v3 source code is freely available on GitHub (https://github.com/klebgenomics/Kaptive), and has been implemented in Kaptive Web (https://kaptive-web.erc.monash.edu). ### Competing Interest Statement The authors have declared no competing interest.
Super excited to finally present the preprint to accompany Kaptive 3 which we released last year!
Big thanks to coauthors @kelwyres.bsky.social, @katholt.bsky.social, @genomarit.bsky.social and Iren Löhr.
Here's what we did to improve in silico antigen typing 👇🧵
www.biorxiv.org/content/10.1...
09.02.2025 03:19 — 👍 21 🔁 12 💬 1 📌 2
Home
A tool for generating consensus long-read assemblies for bacterial genomes - rrwick/Autocycler
New year, new assemblies!
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)
31.12.2024 23:43 — 👍 156 🔁 97 💬 2 📌 3
Please add me
25.11.2024 00:21 — 👍 1 🔁 0 💬 0 📌 0
A small history lesson in #Klebsiella genomics in honour of #KLEBS24…
2009 was a big year for Klebs!
- First genome published
- First description of ST258 KPC-producing clone
pubmed.ncbi.nlm.nih.gov/19218573/
- First description of NDM-1 beta-lactamase
pubmed.ncbi.nlm.nih.gov/19770275/
20.11.2024 13:03 — 👍 48 🔁 22 💬 3 📌 1
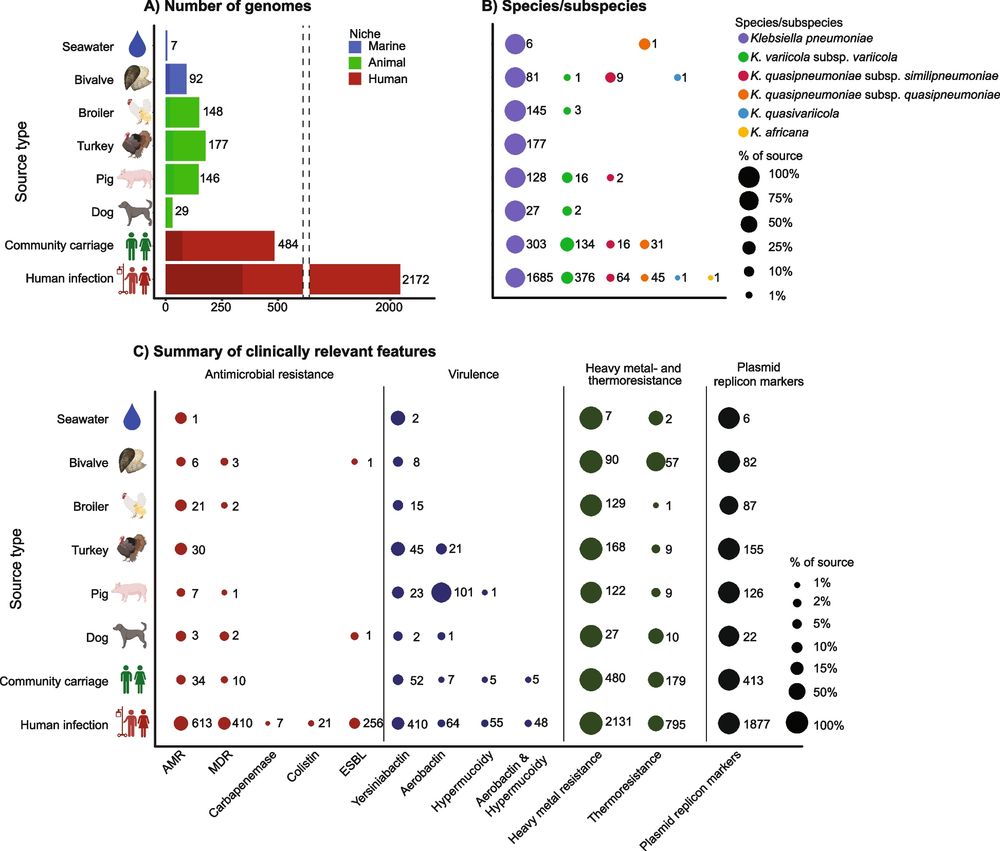
Our study highlights the importance of infection prevention in human clinical settings. However, reducing transmission that leads to colonisation - e.g. from direct contact with animals or via the food chain - could also play an important role in reducing the KpSC disease burden 🦠
13.09.2024 06:20 — 👍 2 🔁 0 💬 1 📌 0

While most KpSC infections in humans were linked to human isolates, we found ~5% had close relatives in animal and marine environments, despite being separated by time and location
13.09.2024 06:20 — 👍 2 🔁 0 💬 1 📌 0
The KpSc populations were distinct but overlapped across human, animal and marine sources, however there were few recent transmission events across the niches
13.09.2024 06:19 — 👍 2 🔁 0 💬 1 📌 0
Excited to share this preprint where we’ve used genomic analysis of >3000 genomes to study Klebsiella pneumoniae in a One Health perspective 🦠🔬👩💻 #OneHealth #KLEBGAP #KlebClub #IrenLöhr @katholt.bsky.social #NORKLEBNET
12.09.2024 16:47 — 👍 36 🔁 22 💬 2 📌 0
PhD studentship with myself and Ed Feil at the University of Bath looking at the evolution and epidemiology of plasmids - in particular looking at plasmid fusions/hybrids/recombinants. Lots of fun bioinformatics! Open to UK or international students.
#microsky
www.findaphd.com/phds/project...
18.10.2023 16:17 — 👍 48 🔁 56 💬 0 📌 2
Postdoctoral researcher at Institut Pasteur | Bioinformatics | Bacterial genomics | Antimicrobial resistance | metagenomics
ESCMID Study Group for mobile elements and plasmids
Publishing reviews and commentaries across the fields of genetics and genomics. Part of Springer Nature and Nature Portfolio.
https://www.nature.com/nrg/
Research fellow in medical microbiology at the University of Liverpool in the UK, studying bacterial genomics, evolution, antimicrobial resistance, and infectious disease epidemiology using bioinformatics and computational biology
🇵🇹 Staff Scientist @probstlab.bsky.social
(@unidue.bsky.social - 🇩🇪). Microbial genomics in the One Health context, biogeochemistry of cave microbiomes, alga-microbe symbioses.
Wellcome Early-Career Fellow @milnerevolution.bsky.social working on mobile genetic element genomics (he/him) 🏳️⚧️🇵🇸
Formerly @biology.ox.ac.uk and @modmedmicro.bsky.social
Assistant Professor & Chancellor Scholar
Department of Microbiology, Biochemistry and Molecular Genetics | Center for Immunity and Inflammation | Rutgers New Jersey Medical School
Host-pathogen interactions, innate immune and metabolic responses
We’re the European Centre for Disease Prevention and Control. We aim to strengthen EU’s defences vs infectious diseases. Here to promote public health.
Bioinformatician @ MDU PHL, Melbourne
Microbial genomics | Computational biology
Located at the Alfred Hospital/Monash Uni
Postdoctoral Research Fellow at the Division of Medical Microbiology, Stellenbosch University, South Africa
Bacteria | Antimicrobial Resistance | Stress Responses | Transcriptomics | Genomics
Repository of new Klebsiella spp. papers posted on Pubmed.
Code maintained by graceeshepard.bksy.social
ESCMID Study Group on Epidemiological Markers
Group leader.
HGT in bacterial pathogens. Acinetobacter, Legionella. Natural transformation. AMR. MGEs.
Centre International de Recherche en Infectiologie
MIBTP Microbiology PhD student
Research Associate, Imperial College London 🍄 Fungal genomics and bioinformatics
Postdoc researcher at Institut Pasteur | Bacterial genomics | Evolution, ecology and diversity of bacterial populations
bacterial genomics・antibiotic resistance https://klebnet.org