
Phased genome assemblies and pangenome graphs of human populations of Japan and Saudi Arabia
The selection of a reference sequence in genome analysis is critical, as it serves as the foundation for all downstream analyses. Recently, the pangenome graph has been proposed as a data model that i...
We made three pangenome graphs 🧬 public, one for the Japanese, one for the Saudi population, and a merged graph (JaSaPaGe). Useful for 🖥️ bioinformatics on either population, or to evaluate how pangenome graphs behave when two different populations are included. jasapage.bio2vec.net/view for PanGene
23.12.2024 09:05 — 👍 7 🔁 2 💬 0 📌 0
Good question; as far as I know, there is no relation between the manuscripts, they are different efforts. Focus is a little different in each manuscript, e.g., in our manuscript we combine a Saudi pangenome graph with a Japanese graph to evaluate effects of combining different populations.
23.12.2024 08:19 — 👍 2 🔁 0 💬 0 📌 0
CAFA 5 Protein Function Prediction
Predict the biological function of a protein
ProtBoost: protein function prediction with Py-Boost and Graph Neural Networks -- CAFA5 top2 solution
The second-place solution in CAFA5 has now been published.
Paper: arxiv.org/abs/2412.045...
GitHub Repo: github.com/btbpanda/CAF...
Kaggle Writeup: www.kaggle.com/competitions...
17.12.2024 22:04 — 👍 5 🔁 3 💬 0 📌 0

A reference quality, fully annotated diploid genome from a Saudi individual - Scientific Data
Scientific Data - A reference quality, fully annotated diploid genome from a Saudi individual
Proud to share our new paper! A complete genome from Saudi Arabia (KSA001), freely available to all. Complex work - not just sequencing & assembly challenges, but also navigating IRB approval to ensure ethical data sharing & open science principles.
nature.com/articles/s41...
24.11.2024 11:12 — 👍 2 🔁 0 💬 0 📌 0
Great work --- would love to be added, will share updates on computational protein function prediction and some complex disease work.
20.11.2024 10:40 — 👍 0 🔁 0 💬 0 📌 0
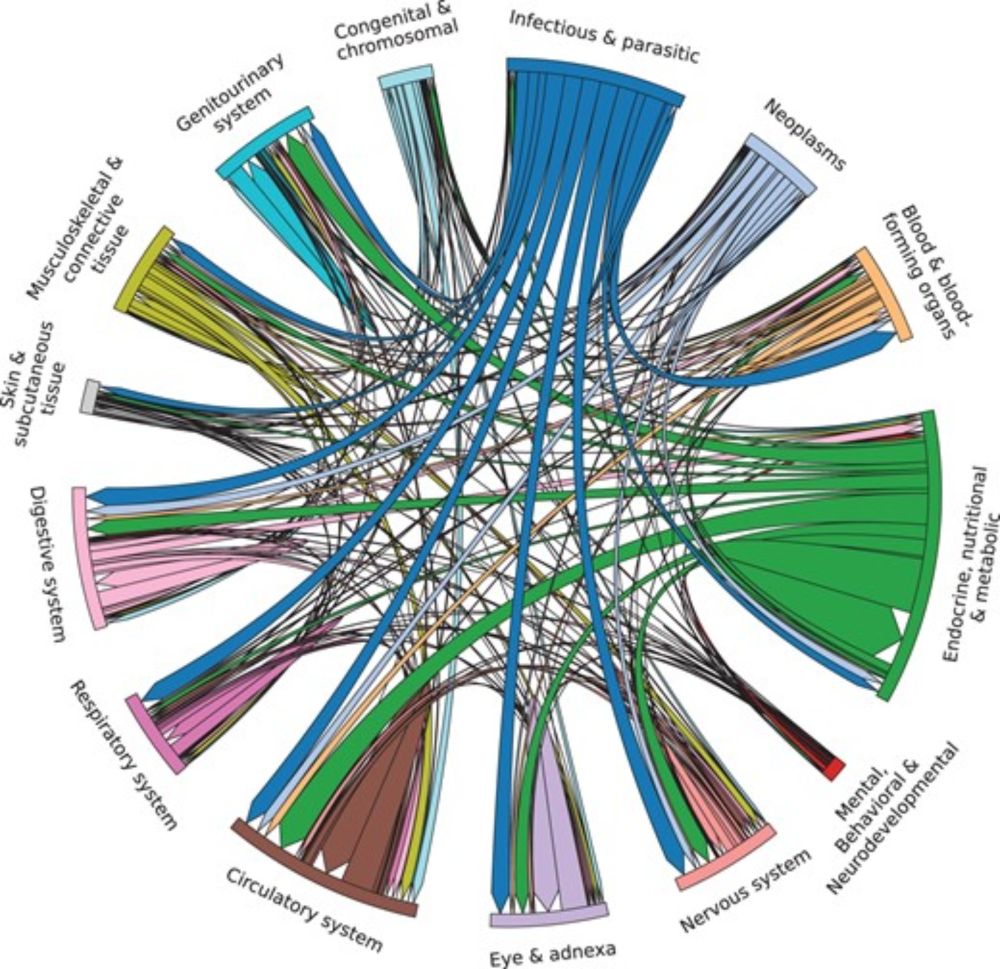
Causal relationships between diseases mined from the literature improve the use of polygenic risk scores
AbstractMotivation. Identifying causal relations between diseases allows for the study of shared pathways, biological mechanisms, and inter-disease risks.
I am excited to share that our paper on creating a very large structure causal model for diseases has been published. We generate an SCM containing most common diseases, and validate with #UKBiobank data, for better polygenic scores, and finding pleitropic variants academic.oup.com/bioinformati...
18.11.2024 04:26 — 👍 10 🔁 1 💬 0 📌 0



