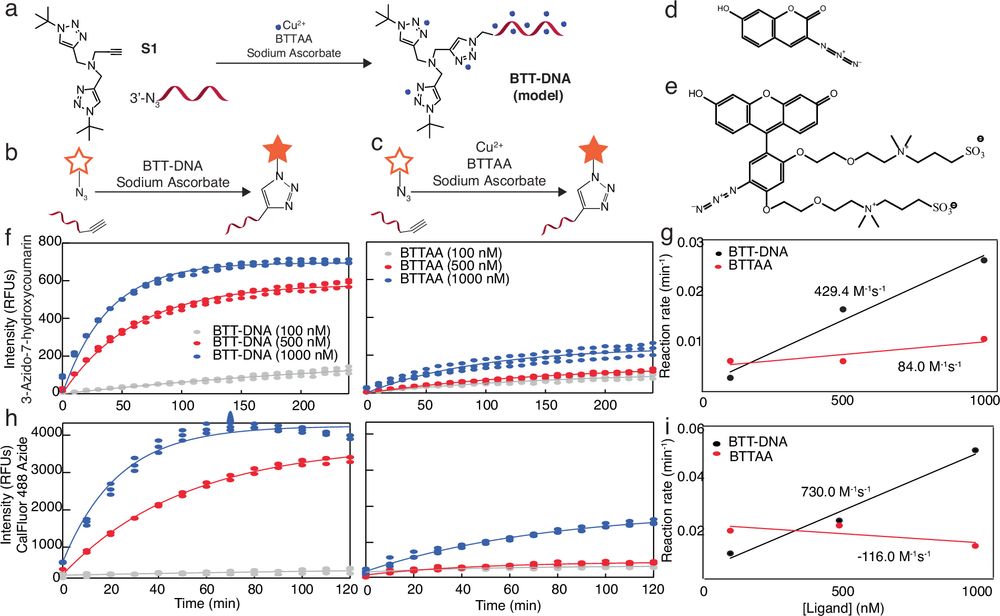
Multi-Scale Kinetics Modeling and Advanced Assay for mRNA-Lipid Nanoparticle Potency Assessment https://www.biorxiv.org/content/10.1101/2025.09.29.679406v1
01.10.2025 23:05 — 👍 0 🔁 1 💬 0 📌 0

RNA modifications in T cells are more stable than expected—primary and immortalized cells share most sites, suggesting common regulatory features can be studied across systems bit.ly/3GP97C8
05.08.2025 19:46 — 👍 1 🔁 1 💬 0 📌 0
Many thanks to the team!! 🤓🤓🤡🥳 @sashafanari.bsky.social @wanunupore.bsky.social
17.07.2025 17:21 — 👍 0 🔁 0 💬 0 📌 0
RNA | Mobile
5/
So are immortalized cells “damaged”? Not quite.
They’re not perfect stand-ins, but they’re surprisingly consistent with primary cells.
We conclude that Jurkat cells are a solid model for studying ψ — just watch out for that 13%.
📖 tinyurl.com/TCellPsiRNA
17.07.2025 17:21 — 👍 0 🔁 0 💬 1 📌 0
4/
The twist? It’s not due to differences in pseudouridine synthase expression — enzyme levels were similar.
So what's driving site selection? Likely trans-regulatory factors or RNA structure, not just enzyme abundance.
17.07.2025 17:21 — 👍 1 🔁 0 💬 1 📌 0

3/ But that other 13%?
☑️ Jurkat-specific ψ-sites hit oncogenic and immune activation genes
☑️ Primary T cell–specific ψ-sites appear on trafficking and calcium signaling genes
🧬 Immortalized cells also showed more clustered ψ patterns, hinting at altered control compared to primary T cells.
17.07.2025 17:21 — 👍 1 🔁 0 💬 1 📌 0

2/ We were genuinely surprised by this result — we expected transformation to cause widespread disruption. But 87% of ψ-sites were shared between Jurkat and primary T cells!
Most differences came from gene expression, not ψ-site selection.
This process seems to be tightly conserved.
17.07.2025 17:21 — 👍 0 🔁 0 💬 1 📌 0
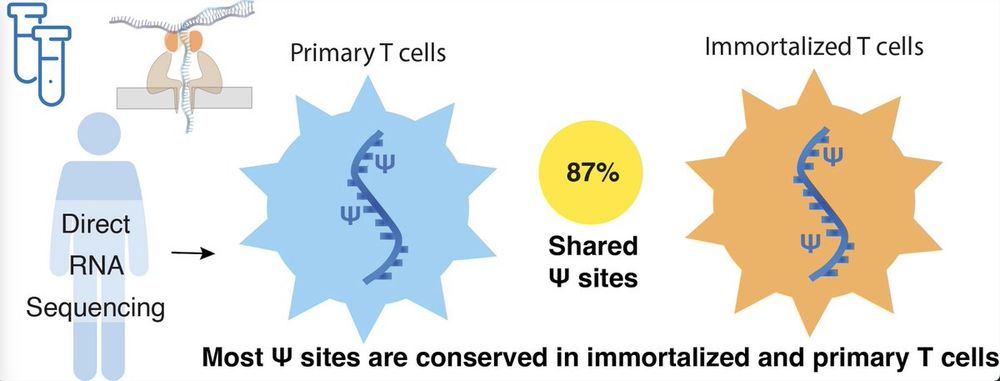
1/🧵
🚨 New paper published in RNA!
Scientists often say anecdotally that RNA modifications are disrupted in immortalized cells — but no one’s really tested it.
So we did: ψ-mapping in primary T cells vs. Jurkat cells using direct RNA-seq.
📄 tinyurl.com/TCellPsiRNA
#nanopore #RNA #pseudouridine
17.07.2025 17:21 — 👍 4 🔁 1 💬 1 📌 0
6/6 🙌 Future work will determine whether static sites are critical for cellular function while plastic sites fine-tune gene expression in response to environmental stressors.
Huge thanks to the team for all their hard work! @wanunupore.bsky.social @genometdcc.bsky.social
21.03.2025 15:42 — 👍 2 🔁 1 💬 0 📌 0
5/6 ⚙️ Enzyme insights: TRUB1 and PUS7 levels shifted with differentiation, and KD experiments reveal unexpected coregulation—knocking down one lowers Ψ at the other’s targets. This hints at a coordinated network fine-tuning mRNA Ψ for neuronal homeostasis and stress resilience.
21.03.2025 15:42 — 👍 0 🔁 0 💬 1 📌 0
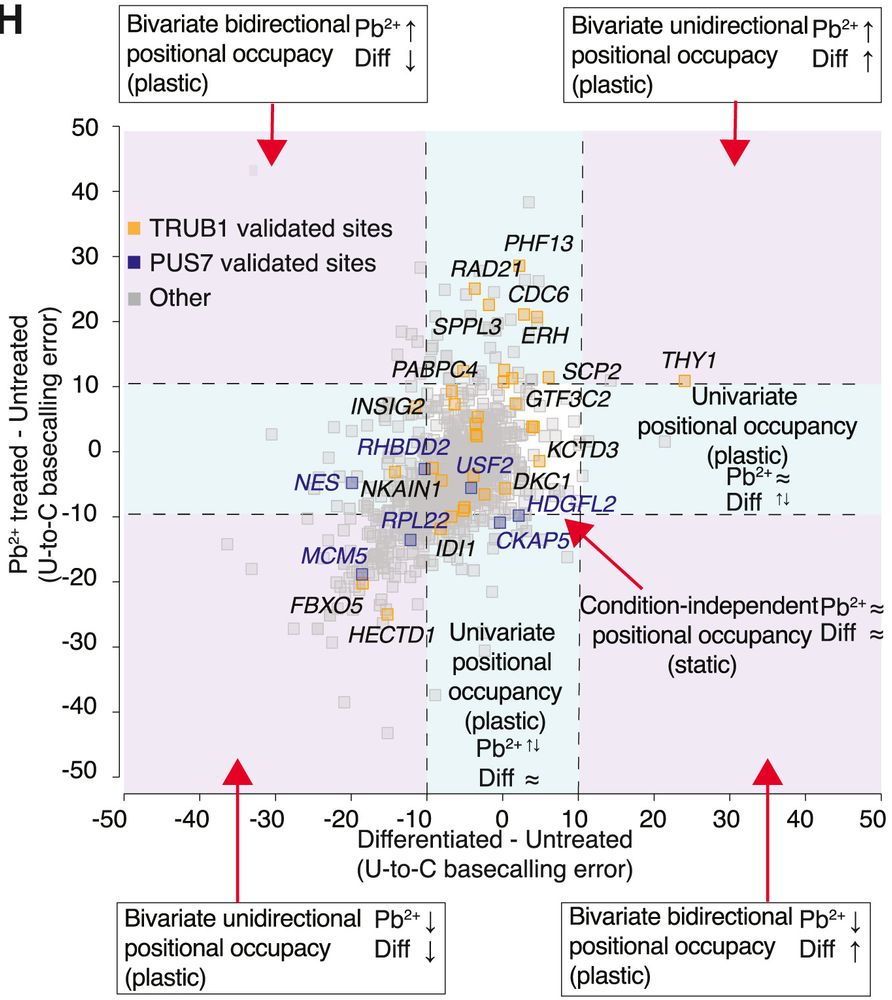
4/6 📊 Plasticity vs stability: ~30% of Ψ sites changed occupancy across conditions (“plastic”), while the rest remained constant (“static”). Among the plastic sites was a key Ψ site in YTHDF1—an m6A reader involved in translation, which changed in response to cellular and environmental cues.
21.03.2025 15:42 — 👍 0 🔁 0 💬 1 📌 0
3/6 📊 Key finding #1: Pb²⁺ treated cells had more Ψ sites transcriptome-wide but lower relative occupancy per site vs untreated/differentiated cells—suggesting a broad but shallow protective pseudouridylation response to stress.
21.03.2025 15:42 — 👍 0 🔁 0 💬 1 📌 0
2/6 🧠Link here: authors.elsevier.com/a/1koBC8YyDf...
Method: SH SY5Y cells were untreated, differentiated with retinoic acid, or exposed to Pb²⁺. Nanopore DRS determined site-specific, relative Ψ “occupancy”; orthogonal knockdowns (TRUB1, PUS7) and biochemical assays validated sites.
21.03.2025 15:42 — 👍 0 🔁 0 💬 1 📌 0

1/6 🧬New paper at @cp-cellsystems.bsky.social led by @sashafanari.bsky.social! Does pseudouridine (Ψ) dynamically respond to cell state? We used nanopore DRS + Mod-p ID to map Ψ in neuron-like cells under “normal” and “perturbed” conditions (healthy differentiation and unhealthy Pb2+ poisoning).
21.03.2025 15:42 — 👍 3 🔁 2 💬 1 📌 1
Sitting in a journal club-style class right now and after the main presenters present, there are "roles" from others in the group, like the "archeologist" who looks into the citation trail, the "private investigator" who tries to dig into the history, and the "industry practitioner", etc. Cool idea!
18.03.2025 15:15 — 👍 57 🔁 13 💬 4 📌 0
Direct RNA sequencing of primary human T cells reveals the impact of immortalization on mRNA pseudouridine modifications. https://www.biorxiv.org/content/10.1101/2025.03.02.641090v1
10.03.2025 17:34 — 👍 5 🔁 3 💬 0 📌 0

Office hours are starting
21.02.2025 14:39 — 👍 2 🔁 0 💬 0 📌 0
I sat on a study section that met this past Friday (2/14)
18.02.2025 01:05 — 👍 2 🔁 0 💬 1 📌 0
Gene editing technology began by people studying salt marshes. Ozempic began by folks studying the venom of Gila Monsters. Support for basic science has empowered us to understand our world. Tethering it to applications health has transformed and saved countless lives.
08.02.2025 13:45 — 👍 3337 🔁 724 💬 49 📌 26
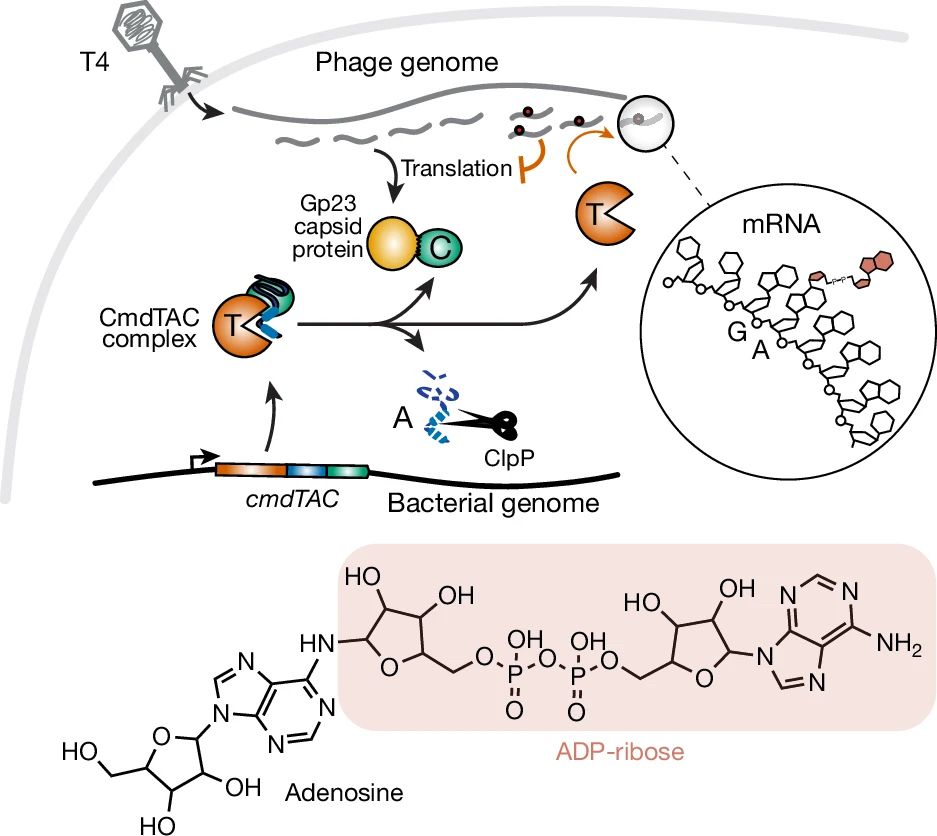
Graphic showing modification of mRNA with ADP-ribose mediated by bacterial enzyme cmdTAC
Check it out! Another novel RNA modification - ADP-ribosylation - that was previously only known on proteins. A cousin to #glycoRNA 👏
www.nature.com/articles/s41...
06.02.2025 19:38 — 👍 223 🔁 81 💬 3 📌 4
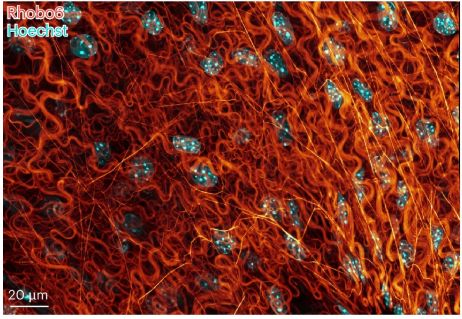
"Imagine a DAPI-like stain, but for the extracellular matrix." That's basically how this work was pitched to me by Kayvon and Antonio a year or so ago. Now the final product really delivers. Read about their versatile label for ECM in living tissues here: www.nature.com/articles/s41...
06.02.2025 16:02 — 👍 354 🔁 104 💬 9 📌 7
Comparative/Evolutionary/Neuro Genetics (he/him) - Personal opinions only
Mosquitoes, watches and whatever. Huck Chair of Disease Epidemiology and Biotechnology at Penn State. My opinions do not reflect those of my employer (they want nothing to do with them whatsoever)
rasgonlab.wordpress.com
Science. Biology. Progress.
Founding Editor of Asimov Press. Subscribe at press.asimov.com!
Colorado RNA biologist & tRNA enthusiast exploring the wild frontiers of nanopore direct RNA sequencing at the intrepid venn diagram of northern blots & machine learning.
Single cell systems and synthetic biology lab at Northwestern University and Chan Zuckerberg Biohub Chicago.
https://www.goyallab.org/
Ecologist, entomologist, writer. Chair of Academic Senate and Professor at CSU Dominguez Hills. ScienceForEveryone.science and I'm the Small Pond Science guy.
he/him
Black Lives Matter. In favor of DEI, justice, access, opportunity. Abolish ICE.
Nature Communications is an open access journal publishing high-quality research in all areas of the biological, physical, chemical, clinical, social, and Earth sciences.
www.nature.com/ncomms/
Co-Founder @Lux Capital | Trustee @SfiScience
Santa Fe Inst | Chair @CiPrep Coney Island Prep (Brooklyn) | Co-Founder of Carson, Quinn & Bodhi w/ @ltwolfe
A monthly, peer-reviewed journal from Cell Press devoted to Systems Biology. Posts from the Editors.
Innate immunity/host-pathogen interactions. Husband, father, former Soviet immigrant. Opinions my own.
Professor and Director, Dept. of Biophysics & Biophysical Chemistry
Johns Hopkins School of Medicine, structural biologist, lover of chromatin and ubiquitin, fan of the active voice.
🏛Senior Lecturer, University of Sheffield 🎓PhD Evolutionary Biology🏝Islands 🐘Elephants🦣Mammoths🦷Teeth ⚒️1/4 of TrowelBlazers 📰EiC Open Quaternary 🎥🎙Presenter. Expcet typos.
Lab Manager @universityofutah.bsky.social @UUNeurobiology | Molecular Mechanisms of Memory | wife and mom of 4 kiddos and a mad dog | views expressed are my own | Lo que escribo es mi opinion.
We aim to serve society by making science open, inclusive, and accessible. Also @500womensci on Instagram and Facebook. Find out more: https://500womenscientists.org/
https://linktr.ee/500womenscientists
science reporter covering biomedical research at Nature | proudly Ukrainian 🇺🇦
maxkozlov.com
signal: mkozlov.01
Drosophila enthusiast. Assistant Professor of Mol Bio and Biochem at Rutgers U. Runner. Uses brain to think about brains. All around curious person.
40 years of NIH grant experience … live jazz & birding keep me sane
Mourning the days when all we talked about was font choice, reference styles and glam humping.
Dare mighty things! Journeyman wondersmith at Speculative Technologies. 🏴☠️🪐🐉
https://Benjaminreinhardt.com + https://spec.tech