
Come and meet us at #EESMicrobiome in Heidelberg! We are excited to share our research with you 🥳
1/3
@annabel-k.bsky.social
PhD student in @tartumicrobiome.bsky.social at the University of Tartu. Interested in early life microbiome development and microbiome-drug interactions | she/they 🌈 | banner photo: Joshua Fuller

Come and meet us at #EESMicrobiome in Heidelberg! We are excited to share our research with you 🥳
1/3
Interested in how it all began? Check out our paper about the Estonian Microbiome Cohort! ➡️ www.nature.com/articles/s41...
06.02.2025 12:05 — 👍 6 🔁 2 💬 0 📌 0
The entire archive of CDC datasets can be found here.
HUGE shoutout to data archivists- this work is important 👏🙌🏻
archive.org/details/2025...

Thank you to organizers of the Research Days of Institute of Biomedicine and Translational Medicine for inviting me to present our project on type 2 diabetes medications and the gut microbiome. I really appreciate the feedback and questions. Find out more from the preprint: doi.org/10.1101/2024...
31.01.2025 13:05 — 👍 12 🔁 1 💬 0 📌 1
Super proud of this paper on infant gut microbiome age coming from my lab. The paper has just been published with @springernature.bsky.social in @naturecomms.bsky.social. Read about it here: rdcu.be/d6cCw.
14.01.2025 15:42 — 👍 52 🔁 12 💬 4 📌 2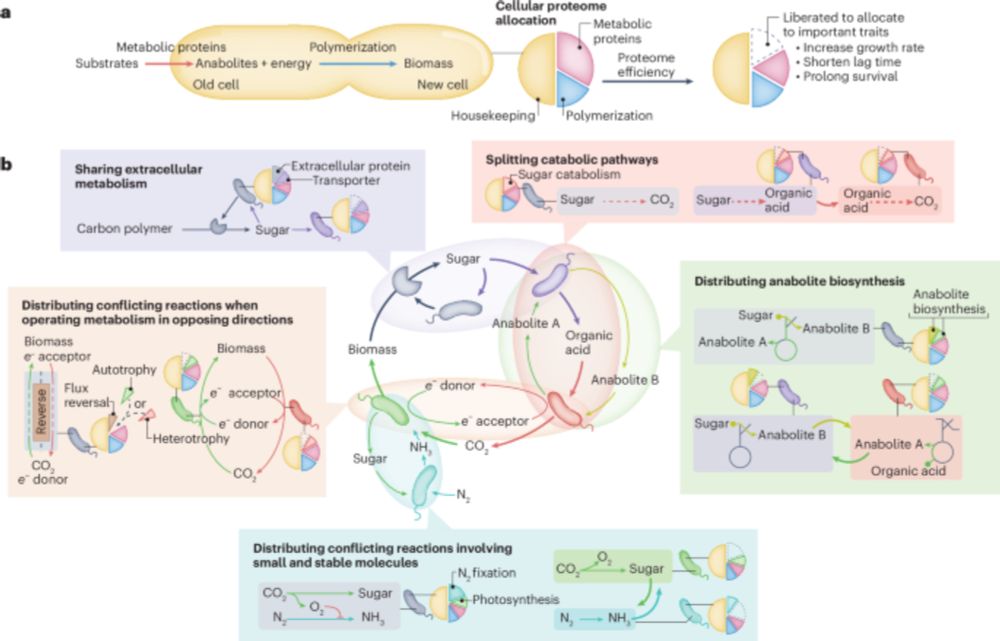
Thrilled to announce that Matthias Hülsmann's new perspective is now out in @naturemicrobiol.bsky.social! This exciting work is shaping how our group thinks about collective microbiome metabolism. Check it out! www.nature.com/articles/s41...
26.11.2024 12:45 — 👍 49 🔁 15 💬 0 📌 0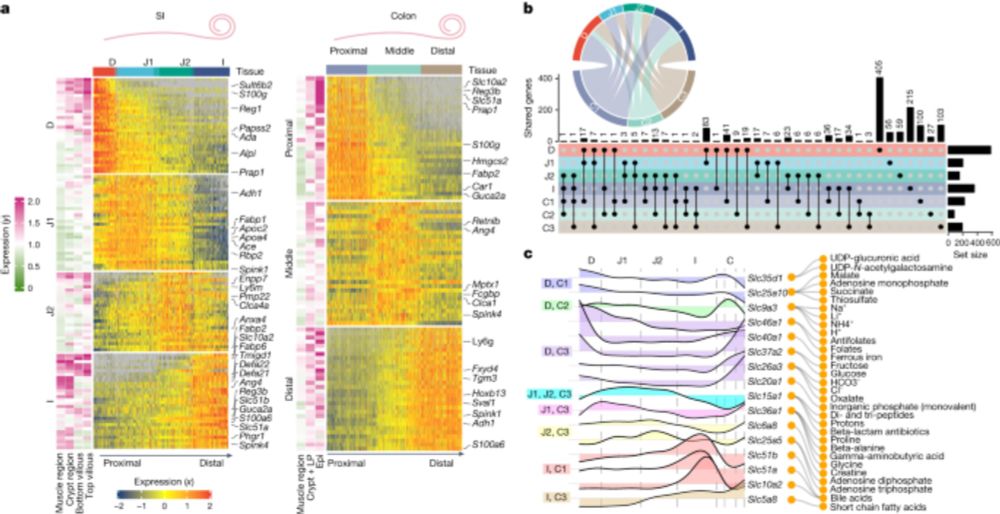
1/ Keen to share the story of my postdoc research out in @natureportfolio.bsky.social! We built a massive resource characterizing the spatial landscape of the gut—and yes, we dug deep into the biology too. If you’ve got a minute, let me walk you through the story. 🧵👇
www.nature.com/articles/s41...
More #NatureReviewsCancer updates 🚨
We recently launched a collection on the #microbiome in cancer. While its role is still debated, the collection presents a selection of articles from #SpringerNature journals to highlight our current understandings.🧫🦠
😀👉 bit.ly/48u8WFH

Your friends shape your microbiome — and so do their friends
Analysis of nearly 2,000 people living in remote villages in Honduras reveals who’s spreading gut microorganisms to whom.
www.nature.com/artic...
1/5


Results 10 yrs metagenomic sequencing for CNS infections www.nature.com/articles/s41...
Impressive.
Key take homes for me.
1) Time to result still long - addressable but a barrier currently.
2) 10% contamination.
3) Despite using cutting edge tests a "causative" organism found in only 15%.

Targeted protein evolution in the gut microbiome by diversity-generating retroelements
www.biorxiv.org/content/10.1...
Nice paper on diversity generating retroelements in the Bacteroides:

Reading: academic.oup.com/nar/advance-...
The mOTUs online database provides web-accessible genomic context to taxonomic profiling of microbial communities

New paper from the lab out in Cell Systems. “Spatiotemporal dynamics during niche remodeling by super-colonizing microbiota in the mammalian gut”
www.cell.com/cell-systems...

Estimates of microbiome heritability across hosts.
#Microbiome #NatureMicrobiology #BioSky #SciSky 🧪
www.nature.com/articles/s41...
Hi, Jan! I would love to be added there. Thank you!
15.11.2024 08:04 — 👍 1 🔁 0 💬 1 📌 0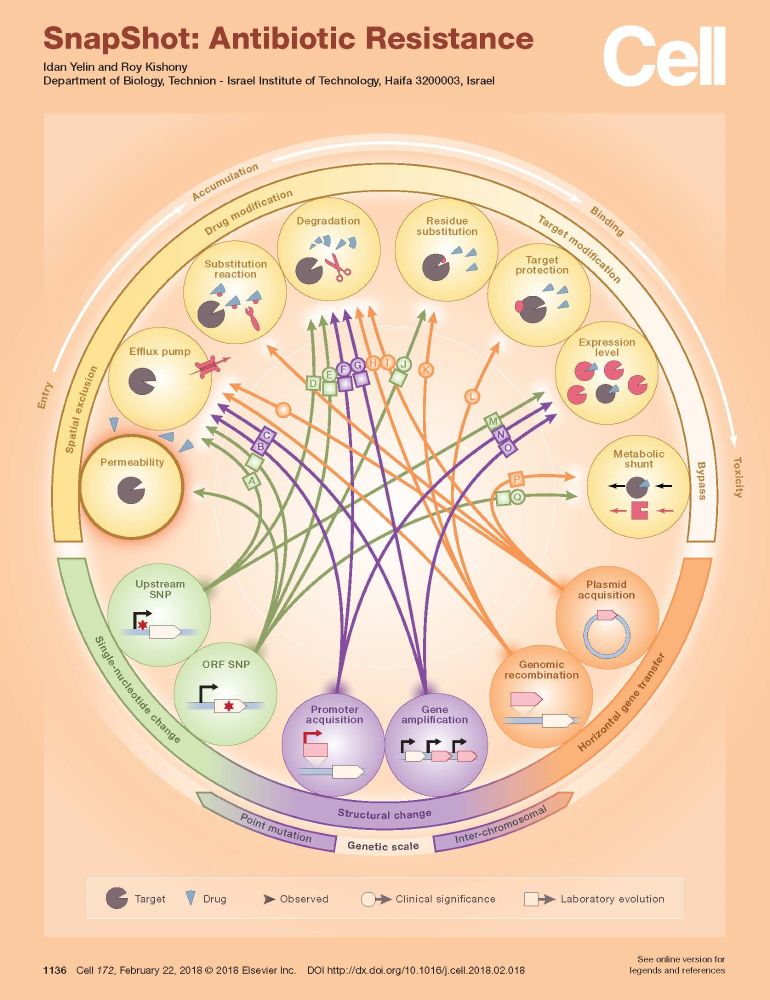
Incredible graphic - the myriad of ways that bacteria defend themselves against antibiotics.
14 resistance mechanisms, summarised by Idan Yelin & Roy Kishony in Cell
www.sciencedirect.com/science/arti...
Hot off the press: "Fecal microbial load is a major determinant of gut microbiome variation and a confounder for disease associations" www.sciencedirect.com/science/arti...
See below for @suguru-nishijima.bsky.social 's thread on the preprint!
#microsky #microbiome
Excited to share our latest preprint on a novel computational tool to predict fecal microbial load solely from relative species profiles! Our study revealed that fecal microbial load is a substantial confounder in microbiome-disease association studies.
www.biorxiv.org/content/10.1...

Bluesky, I want to share with you one of my favourite papers:
09.11.2024 16:23 — 👍 102 🔁 31 💬 4 📌 3
After editing, this very short letter to New Scientist is even shorter now. But I think it gets the point across! @faecalmatters.bsky.social
11.11.2024 22:23 — 👍 120 🔁 34 💬 10 📌 4
Your hitchhiker's guide to microbiome studies:
journals.asm.org/doi/epub/10....

I'm new here, so I'll share our recent work here too 😁🧫
Using Tn-seq + live imaging in engineered airway organoids, we explored the fitness tradeoffs P. aeruginosa faces while growing and surviving antibiotic treatment on the mucosal surface.
Check it out: www.nature.com/articles/s41...
#MicroSky
Welcome all (new) followers! Besides our work on probes & biosensors I'd like to share what I know about #Rstats and #DataViz. So I composed a book on data visualization with R & ggplot2 (it's work in progress).
Target audience = wet lab scientists.
Link: joachimgoedhart.github.io/DataViz-prot...
[looking at Bluesky over the past 2 days] So this is what the Cambrian explosion must have felt like
11.11.2024 22:17 — 👍 957 🔁 186 💬 26 📌 13
Human xenobiotic metabolism proteins have full-length and split homologs in the gut microbiome https://www.biorxiv.org/content/10.1101/2024.11.06.622278v1
08.11.2024 22:34 — 👍 1 🔁 1 💬 0 📌 0
Hello, Bluesky! I'm Annabel, a PhD student in the Tartu Microbiome group led by Prof. Elin Org at the University of Tartu (EE).
I am interested in the long-term effects of medications on the gut microbiome and early-life microbiome development.
I also like photography, sci-fi, and crocheting 🌿