Mercredi prochain je serai de retour à Villejuif pour les 10 ans d’iGEM IONIS.
Table ronde sur la biologie de synthèse face aux enjeux climatiques que je partagerai avec Andrew Tolonen… mon prof en 2015 et advisor IGEM en 2014.
18.09.2025 19:32 — 👍 0 🔁 0 💬 0 📌 0
Excited to announce a preprint describing our software package Meeko! Meeko is a Python package that uses RDKit for receptor and ligand preparation, including protonation, bond order, and connectivity and processing of docking results. It is customizable and suitable for high-throughput workflows.
18.09.2025 00:37 — 👍 3 🔁 2 💬 1 📌 0

New Practical Cheminformatics Post
patwalters.github.io/Three-Papers...
22.07.2025 13:40 — 👍 16 🔁 9 💬 0 📌 2
With the release of Boltz 2, is there already a review comparing the efficiency of different (new) docking tools? (I’ve already tried Pocket Vina, which is actually quite good for high-throughput)
31.08.2025 12:08 — 👍 3 🔁 1 💬 0 📌 1

Entre deux simulations, épris de rationalité et mu par le désir de progrès, je me plonge dans cet ouvrage qui saura sans doute tempérer mes ardeurs…
31.08.2025 09:41 — 👍 2 🔁 1 💬 0 📌 0
I'm excited to share our new preprint: PocketVina — a fast, scalable, and accurate multi-pocket molecular docking method.
Docking remains essential in early-stage drug discovery, but recent deep learning–based approaches still face limitations in generating...
Thread - (1/n)
26.06.2025 11:11 — 👍 4 🔁 4 💬 1 📌 1
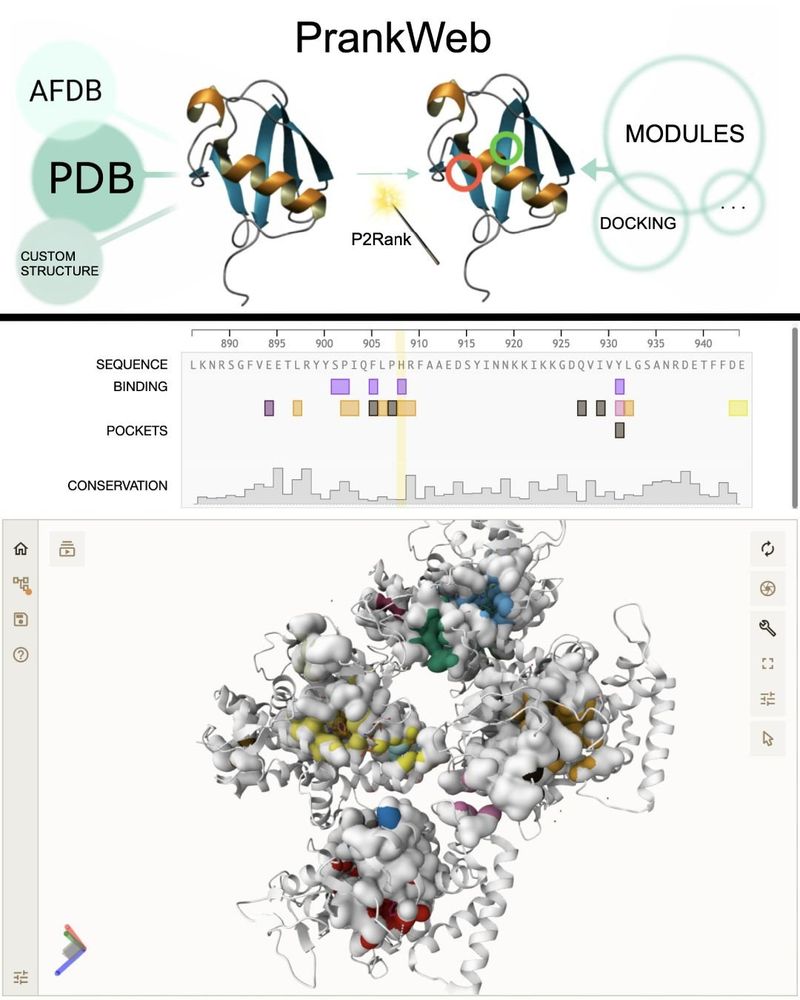
Screenshot of PrankWeb.
🚀 Our paper on the updated PrankWeb, a web app for protein binding site prediction using P2Rank, is out in
@narjournal.bsky.social. Now with docking integration!
👨💻 Try it: prankweb.cz
📄 Paper: doi.org/10.1093/nar/...
💻 GitHub: github.com/cusbg/prankweb
Developed at @cusbg.bsky.social
10.06.2025 15:09 — 👍 24 🔁 7 💬 2 📌 2

The heptagonal box model is a specificity and activity model that helps identify the type of reaction an enzyme catalyzes and its substrate, but it is not meant to indicate how well the enzyme performs. Structural features clearly play a critical role in catalysis (dynamics, binding, etc.).
24.05.2025 11:56 — 👍 2 🔁 0 💬 0 📌 0
It actually feels like the opposite here in Paris. I keep seeing more and more people with a book in hand on public transport. Even at Abolis, there's a growing habit of sharing and recommending books.
06.05.2025 12:31 — 👍 1 🔁 0 💬 1 📌 0
Close to recreating it in ChimeraX:
1. Install the python file: github.com/smsaladi/chi...
2. rainbow palette magma
3. col modify #1 blackness - 40
4. col modify #1 whiteness - 40
5. col modify #1 saturation - 50
6. col modify #1 lightness + 20
Room for improvement, needs more desaturation
16.04.2025 06:31 — 👍 53 🔁 9 💬 3 📌 1
Introducing CAMEO Structures & Complexes - automated weekly blind benchmarking of structure prediction servers. Now with heteromeric and protein-ligand complexes.
Join us and register your server now!
cameo3d.org
28.04.2025 07:56 — 👍 7 🔁 7 💬 1 📌 3
"De novo prediction of protein structural dynamics"
I'll be presenting an overview of the field tomorrow at a workshop. Link to a PDF copy of the presentation: delalamo.xyz/assets/post_...
27.04.2025 14:16 — 👍 70 🔁 17 💬 5 📌 1

AFESM: a metagenomic guide through the protein structure universe! We clustered 821M structures (AFDB&ESMatlas) into 5.12M groups; revealing biome-specific groups, only 1 new fold even after AlphaFold2 re-prediction & many novel domain combos. 🧵
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...
27.04.2025 00:13 — 👍 141 🔁 71 💬 4 📌 4
Gene synthesis is often the most expensive part of protein engineering with generative models.
Happy to have played a small part in this work, where Chase developed a method for precision library construction at scale, with per-gene costs as low as $1.50.
@philromero.bsky.social
24.03.2025 17:24 — 👍 63 🔁 23 💬 1 📌 0
Three BioML starter packs now!
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
Pack 3: go.bsky.app/NAKYUok
DM if you want to be included (or nominate people who should be!)
03.12.2024 03:27 — 👍 147 🔁 59 💬 16 📌 6
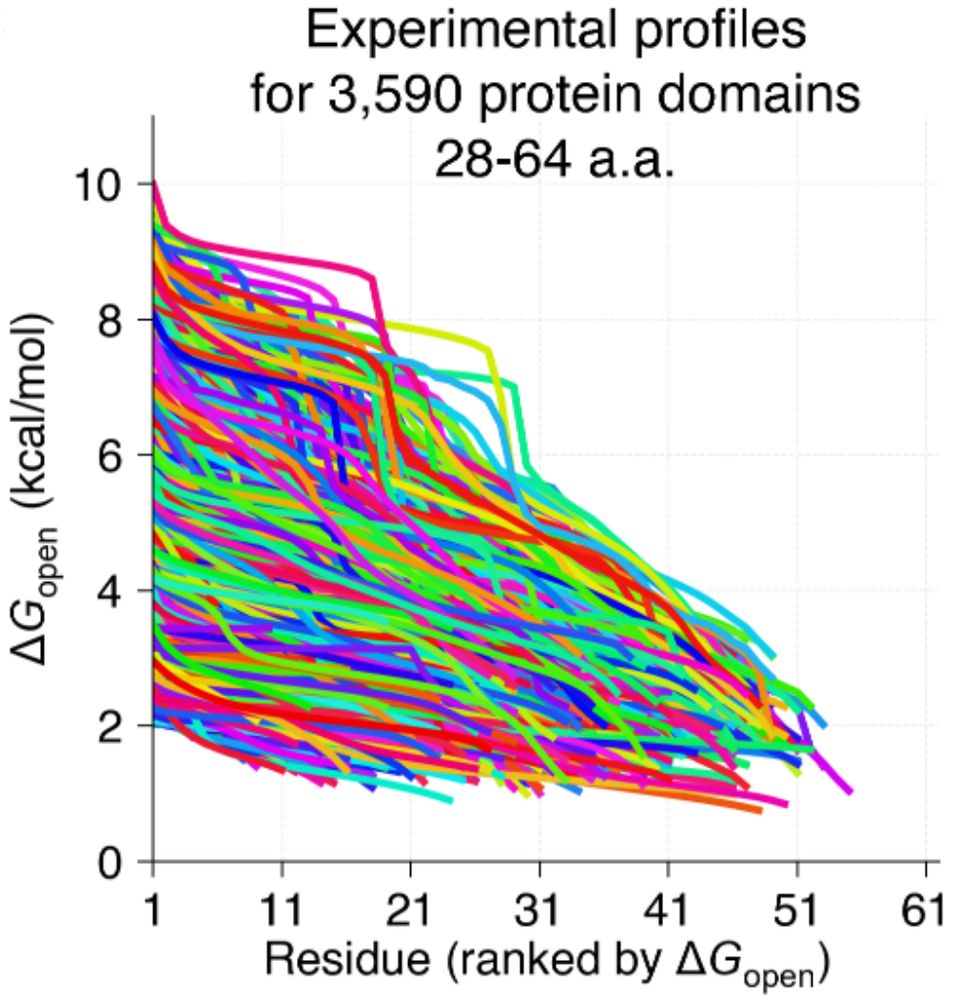
Small proteins can be more complex than they look!
We know proteins fluctuate between different conformations- but by how much? How does it vary from protein to protein? Can highly stable domains have low stability segments? @ajrferrari.bsky.social experimentally tested >5,000 domains to find out!
26.03.2025 16:21 — 👍 87 🔁 36 💬 4 📌 0

For future reference, this is the slide (without the bottom cropped) that should have been posted here. Boltz-1 and AF3 are actually performing similarly on the Chymase and Mpro datasets, and differ only on the Autotaxin dataset for still unknown reasons.
09.12.2024 15:38 — 👍 16 🔁 6 💬 0 📌 0

GitHub - baker-laboratory/PLACER: PLACER is graph neural network for local prediction of protein-ligand conformational ensembles.
PLACER is graph neural network for local prediction of protein-ligand conformational ensembles. - baker-laboratory/PLACER
We are happy to share that the code and weights of PLACER (formerly known as ChemNet) are now public!
This method allows for rapid evaluation of protein side chain and ligand conformational ensembles, with uses in ligand docking and enzyme evaluation.
github.com/baker-labora...
13.02.2025 20:28 — 👍 30 🔁 11 💬 1 📌 1
Rediriger la technologie au service de l'intérêt général 💪
https://dataforgood.fr
Toutes les manières d'éclairer le monde grâce à la philosophie.
https://www.philomag.com/
Cheminformatics, ML, Drug Discovery
Computational Structural Biology @biozentrum.unibas.ch @unibas.ch @sib.swiss | President of the Research Council of the @snsf-ch.bsky.social | Team Science 🧪
PI at Institut Pasteur
Evolution, immunity, genomics, microbiolgy.
Into immunity in bacteria and its conservation in eukaryotes.
Advocate for more inclusive sciences
https://research.pasteur.fr/en/team/molecular-diversity-of-microbes/
Média qui alerte sur un truc pas cool : le changement climatique
Nous soutenir : https://bonpote.com/soutenir
https://bonpote.com/
Journaliste climat @mediapart.fr • Auteur de "Le Mensonge Total" (2024), de "Criminels climatiques" (2022) et de "Une Histoire Populaire du Football" (2018).
Pour une écologie politique qui se prend au sérieux, mêlant les sciences, les luttes progressistes et révolutionnaires, et la philosophie.
Venez sur twitch pour la revue hebdomadaire, le matin tous les weekends.
https://www.twitch.tv/mif_934
Group Leader @ Institut Curie. Working on geometric deep learning for protein and RNA structures representation. Interested in drug design applications.
Protein dynamics and protein design. But so far only with a computer... Yes I'm happy to rely on excellent collaborators who do the dirty work! :-)
Professor of Bioinformatics at the University of Reading, developer of methods for protein structure and function prediction.
https://www.reading.ac.uk/bioinf/
Machine Learning and Computational Biology at Helmholtz Munich, UZH, ETH Zurich, and previously at Genentech
Working as Head of Talent and Culture at Xyme.ai.
'Enzyme design is the next grand challenge. Nature has spent billions of years evolving the enzymes we use today. Now we must design the enzymes of tomorrow.'
https://jobs.ashbyhq.com/xyme
computation biology, drug discovery, computer vision, microscopy, statistics, machine learning, all happening at https://carpenter-singh-lab.broadinstitute.org/
Senior Chemøinformatician at Merck | Postdoc@Broad Institute of MIT and Harvard, Cambridge(MA) | PhD@University of Cambridge(UK) | AI, Image Analysis, bioML, -omics data, and Cell Painting for drug discovery. srijitseal.com/tools
https://www.rdkit.org
https://github.com/rdkit/rdkit
A bit of maths, a bit of blockchain, books, and some stuff about computer programming
Computational biologist @biozentrum.bsky.social. Likes protein structures.
https://ninjani.github.io/
We create proteins that solve modern challenges in medicine, technology, and sustainability.
• 2024 Nobel Prize in Chemistry
• University of Washington, Seattle
→ ipd.uw.edu