Image of crowded bookshelves.
A lot is changing with our documentation, including a transition from a single-source to multiple sources. Learn more in this News post on the forum: forum.qiime2.org/t/the-qiime-...
#qiime2 #bioinformatics #documentation #jupyterbook
02.07.2025 12:42 — 👍 0 🔁 0 💬 0 📌 1
QIIME 2 2024.2 is now available!
The QIIME 2 2024.2 release is now available! Thanks to everyone involved for their hard work! 🙌🏼 🎉 As a reminder, our next planned QIIME 2 release is scheduled for May 2023 (QIIME 2 2023.5), but plea...
Happy Monday! Start your week off w/ a new QIIME 2 release: @qiime2 2024.2. There's a lot in this one, including new parallel actions in the shotgun distribution, a fancy new visualizer (summarize-plus), for summarizing feature tables & performance enhancements and minor fixes throughout. Enjoy!
19.02.2024 18:16 — 👍 0 🔁 1 💬 0 📌 0
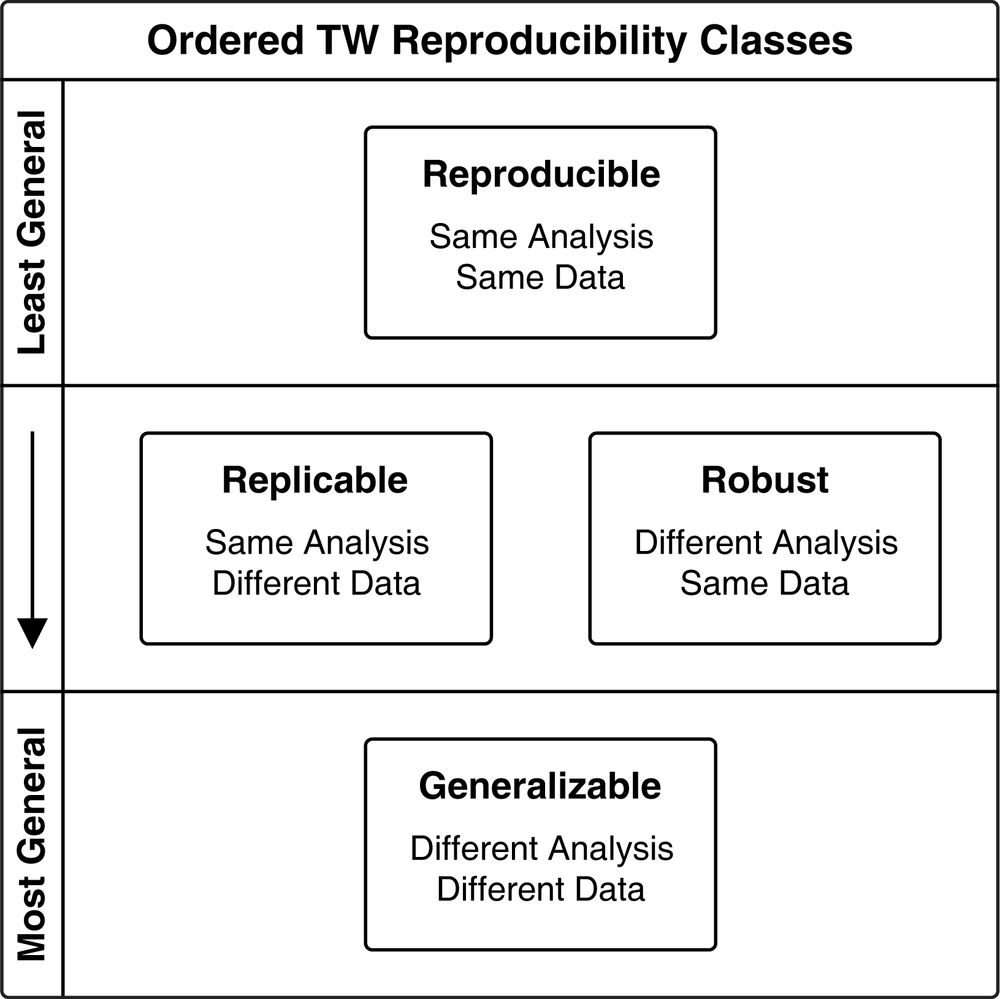
Definitions of Reproducible, Replicable, Robust, and Generalizable in the context of data analysis.
QIIME 2 Provenance Replay auto-generates the code for creating QIIME 2 results from QIIME 2 results & builds "bioinformatics reproducibility supplements" for your papers. Our goal is to make it easy for you to perform reproducible, replicable & robust bioinformatics.
doi.org/10.1371/jour...
29.11.2023 15:51 — 👍 3 🔁 1 💬 0 📌 2
#QIIME2 2023.9 is now available! Exciting updates include an alpha release of our shotgun #metagenomics distribution, fully integrated provenance replay within the framework, batched decontam analysis, and more! Thank you contributors!!
forum.qiime2.org/t/qiime-2-20...
20.10.2023 18:49 — 👍 2 🔁 1 💬 0 📌 0