
Next Tues (6/3) at 4PM ET, we will have @avapamini.bsky.social
present "Deep learning guided design of protease substrates"
Paper: www.biorxiv.org/content/10.1...
Sign up on our website for zoom links!
@ml4proteins.bsky.social
Seminar series on machine learning for protein engineering. Details for seminars and slack are on our website. DM us if you're interested in presenting! ml4proteinengineering.com

Next Tues (6/3) at 4PM ET, we will have @avapamini.bsky.social
present "Deep learning guided design of protease substrates"
Paper: www.biorxiv.org/content/10.1...
Sign up on our website for zoom links!

Next Tues (5/20) at **2PM ET**, we will have Woody Ahern and Jason Yim present "Atom level enzyme active site scaffolding using RFdiffusion2"
Paper: www.biorxiv.org/content/10.1...
Sign up on our website for zoom links!

Next Tues (5/13) at 4PM ET, we will have Anna Lauko and Sam Pellock present "Computational design of serine hydrolases"
Paper: www.science.org/doi/10.1126/...
Sign up on our website for zoom links!

Next Tues (4/29) at **4:30PM** ET, we will have @ginaelnesr.bsky.social @hkws.bsky.social present "Learning millisecond protein dynamics from what is missing in NMR spectra"
Paper: biorxiv.org/content/10.1...
Sign up on our website for zoom links!
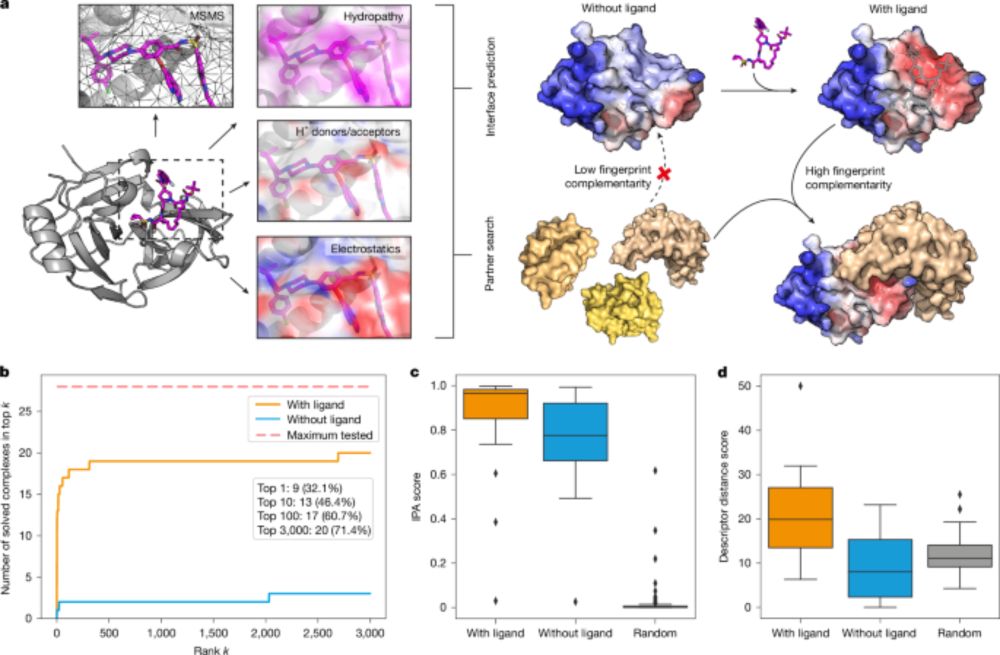
Next Tues (4/1) at **11AM** ET we will have Anthony Marchand present "Targeting protein–ligand neosurfaces with a generalizable deep learning tool"
Paper: www.nature.com/articles/s41...
Sign up on our website for zoom links!

Next Tues (3/18) at 4PM ET we will have Alan Amin
present "Bayesian Optimization of Antibodies Informed by a Generative Model of Evolving Sequences"
Paper: arxiv.org/abs/2412.07763
Sign up on our website for zoom links!

Next Tues (3/4) at 4PM ET we will have Karsten Kreis present "Proteina: Scaling Flow-based Protein Structure Generative Models"
Paper: openreview.net/forum?id=TVQ...
Sign up on our website for zoom links!

Next Tues (2/18) at **5PM ET** we will have @hannes-stark.bsky.social and Bowen Jing present "ProtComposer: Compositional Protein Structure Generation with 3D Ellipsoids"
Paper: openreview.net/forum?id=0ct...
Sign up on our website for zoom links!

Next Tuesday, 2/4 @ 4 pm EST, Aya Ismail and Nathan Frey will present "Concept Bottleneck Language Models"
Paper: arxiv.org/abs/2411.06090
Sign up on our website to receive Zoom links!

Next Tuesday, 1/21 @ 4 pm EST, Elana Simon
will present "InterPLM: Discovering Interpretable Features in Protein Language Models via Sparse Autoencoders"
Paper: www.biorxiv.org/content/10.1...
Sign up on our website to receive Zoom links!

This Tuesday, 1/7 @ 4 pm EST, we'll be back with the 2025 seminar series!
Seyone Chithrananda will present "Mapping the combinatorial coding between olfactory receptors and perception with deep learning"
Paper: biorxiv.org/content/10.1...
Sign up on our website to receive Zoom links!