Huge shoutout to the amazing instructor squad @kaplipa.bsky.social , @sabifo4.bsky.social , Tomas Flouri & @zihengyang.bsky.social for smashing another epic #Phylogenomics course! 🚀
Massive thanks to everyone who joined us this week — go crush your data and rock those projects! 💥🔥
05.12.2025 15:11 — 👍 6 🔁 3 💬 0 📌 1

Ziheng Yang's and Mario dos Reis' research groups, and visitors, at the Grant Museum of Zoology at University College London. Left to right: Jiayi Ji, Chi Zhang, Yuttapong Thawornwattana, Lucas Freitas, Zihui Yang, Ziheng Yang, Yuqing Peng (bottom), Siri Olsen (top), Laura Mulvey, Mario dos Reis (bottom), Asif Tamuri (top), Sandra Álvarez-Carretero.
Wrapping up our annual joint lab meeting with a visit to the Grant Museum of Zoology at UCL was such a highlight! Always inspiring to reconnect with longtime colleagues, new team members, and collaborators💫
25.11.2025 10:12 — 👍 8 🔁 1 💬 0 📌 0

Workshop on Computational Genomics at Beijing Normal University (Nov 6th - 7th 2025) | Ziheng Yang focusing on the Gamma prior during the BPP sessions

Workshop on Computational Genomics at Beijing Normal University (Nov 6th - 7th 2025) | Sandra Álvarez-Carretero teaching Bayesian inference using MCMC

Workshop on Computational Genomics at Beijing Normal University (Nov 6th - 7th 2025) | Xiyun Jiao teaching gene flow under the MSc

Workshop on Computational Genomics at Beijing Normal University (Nov 6th - 7th 2025) | Tomas Flouri teaching models of sequence evolution and maximum likelihood
And some more pictures from the workshop! (2/2) Thanks to lab members @sabifo4.bsky.social , Tomas Flouri, and Yuttapong Thawornwattana!
11.11.2025 06:09 — 👍 4 🔁 0 💬 0 📌 0

Workshop on Computational Genomics at Beijing Normal University (Nov 6th - 7th 2025) | Group picture

Workshop on Computational Genomics at Beijing Normal University (Nov 6th - 7th 2025) | Co-organisers Ziheng Yang and Dayong Zhang with participant and course assistant Cai-jin Chen

Workshop on Computational Genomics at Beijing Normal University (Nov 6th - 7th 2025) | Introductions

Workshop on Computational Genomics at Beijing Normal University (Nov 6th - 7th 2025) | From left to right: Sirui Cheng (instructor), Yuttapong Thawornwattana (instructor), Tomas Flouri (instructor), Jie Han (co-organiser), Sandra Álvarez-Carretero (instructor), Xiyun Jiao (instructor), Ziheng Yang (co-organiser and instructor)
We have had a great time at the Workshop on Computational Genomics at Beijing Normal University! Thank you to all organisers, course assistants, and the fantastic and hard-working participants for making this event possible! 下次见, 谢谢 🌹(1/2)
11.11.2025 06:04 — 👍 5 🔁 0 💬 1 📌 1

On measures of influence and discordance in phylogenomic analyses
Abstract. As models are approximate descriptions of real biological processes, and data are collected often with errors and contamination, sensitivity or r
First up we have Xiyun Jiao and @zihengyang.bsky.social showing the importance of moving beyond model fit testing and embracing sensitivity testing in phylogenetics.
In other words, question your data assumptions and test for model robustness not just adequacy.
(2/n)
doi.org/10.1093/evol...
29.09.2025 16:00 — 👍 3 🔁 1 💬 2 📌 0

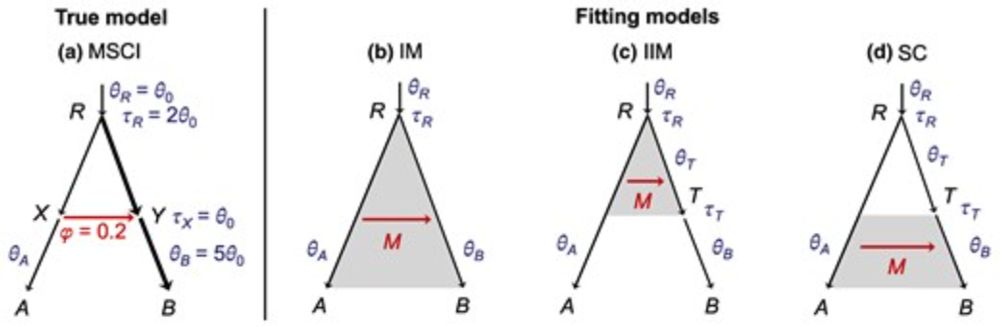
Ziheng Yang, SMBE2025 - Six impossible things to believe before breakfast, about gene flow

Ziheng Yang, SMBE2025 - Six impossible things to believe before breakfast, about gene flow
Ziheng Yang, SMBE2025 - Six impossible things to believe before breakfast, about gene flow

Ziheng Yang, SMBE2025 - Six impossible things to believe before breakfast, about gene flow
In the first Plenary speech of #SMBE2025, Ziheng Yang (@zihengyang.bsky.social) taught us quite a few improbable things on gene flow.
We wish you a delicious breakfast for tomorrow, while mulling over these lessons... 😉
20.07.2025 16:18 — 👍 11 🔁 2 💬 0 📌 0

The first Keynote speech of #SMBE2025 will be given by SMBE's President Ziheng Yang (@zihengyang.bsky.social).
"Six impossible things to believe before breakfast, about gene flow"
20.07.2025 | 18h00
23.06.2025 09:02 — 👍 24 🔁 5 💬 1 📌 1

Organisers of the 2025 Computational Molecular Evolution course. From left to right: Cilia Antoniou, Alexis Stamatakis, Adam Leaché, and Ziheng Yang

Lunch at a Cretan taverna after the social hike.

CoME25 closed the karaoke session at the hotel where we were staying!

Some of the participants and instructors at the lunch break at the beach.
And... Some more pictures from the social activities! There was even time to enjoy the beach during lunch breaks and group singing at an evening karaoke session! 🎤🏖️
06.06.2025 14:31 — 👍 2 🔁 1 💬 0 📌 0

Instructors and participants at the 2025 Computational Molecular Evolution course in Crete (group picture).

Sandy teaching Bayesian node-dating analyses for timetree inference at the Computational Molecular Evolution course.

Attendees joining the 2025 Satellite Workshop "Biodiversity Genomics: sample types, data, and tools to assess past, present and future biodiversity" in Crete (group picture)

Ziheng's talk at the satellite workshop "Biodiversity Genomics: sample types, data, and tools to assess past, present and future biodiversity". Talk title: "Inference of between-species gene flow using genomic data"
What a busy (but sunny) May in Crete!🔆 Ziheng & @sabifo4.bsky.social enjoyed teaching and meeting the participants at the Computational Molecular Evolution course! Ziheng then joined the satellite workshop on Biodiversity Genomics 🌍🧬 Thanks to everyone who made these fantastic meetings possible!!
06.06.2025 14:15 — 👍 8 🔁 1 💬 1 📌 1

Post viva celebrations with Dr Jiayi Yi. Left to right: Hernán Burbano, Jiayi Ji, Simon Martins, and Ziheng Yang.

Post viva celebrations with Dr Jiayi Yi. Jiayi holding one of his PhD presents: a PhD mug and a cute chipmunk plush.

Post viva celebrations with Dr Jiayi Yi. Jiayi showing his new chipmunk plush.
Congratulations Dr Jiayi Ji!! 🥳Thanks to examiners @hernanaburbano.bsky.social and Simon Martin, now time to enjoy the long weekend! 🎉
02.05.2025 20:06 — 👍 7 🔁 2 💬 1 📌 0

And that's a wrap on the SystAssn-SRUK/CERU Bayesian Phylogenetics Workshop!
Thank you to all the attendees, instructors (@anaserrasilva.bsky.social, @sabifo4.bsky.social, Tomas Flouri and Yuttapong Thawornwattana) and to our wonderful TA @lauramulvey.bsky.social!
30.04.2025 18:22 — 👍 7 🔁 2 💬 0 📌 1
The 2nd day of the SystAssn-SRUK/CERU Bayesian Phylogenetics Workshop was focused on the Multispecies Coalescent Theory and the usage of BPP, led by our lab members Tomas Flouri and Yuttapong Thawornwattana! 💻🧬
01.05.2025 09:05 — 👍 2 🔁 1 💬 0 📌 0
The SystAssn-SRUK/CERU Bayesian Phylogenetics Workshop started yesterday! During the 1st day, our lab members Tomas Flouri, @anaserrasilva.bsky.social, and @sabifo4.bsky.social had fun sharing their knowledge on Bayesian statistics, phylogeny inference & timetree inference with the participants💻🧬
30.04.2025 11:15 — 👍 4 🔁 1 💬 0 📌 0
Our lab members @sabifo4.bsky.social, @anaserrasilva.bsky.social, Tomas Flouri, and Yuttapong Thawornwattana are teaching a two-day workshop on Bayesian Phylogenetics Workshop 💻🧬 Pre-register your interest to join this event before March 14th, do not miss the deadline! More details below 🔽
19.02.2025 17:49 — 👍 2 🔁 2 💬 0 📌 0
Thinking of starting a PhD on Bayesian phylogenetics next October 2025 in London? 🎓
You may want to check the TREES project led by the dos Reis Lab on integrating morphology and genomes for timetree inference! 🦴🧬💻Project supervisors: @mariodosreis.bsky.social & Ziheng Yang. More info in link 🔽
09.12.2024 12:09 — 👍 9 🔁 6 💬 1 📌 1

Wrapping up an incredible 5-day course on #Phylogenomics with a fascinating lecture on Molecular-Clock dating of species divergence by @mariodosreis.bsky.social, followed by a hands-on session with @sabifo4.bsky.social 🧬🕰️
Huge thanks to all participants for an inspiring week!@zihengyang.bsky.social
06.12.2024 11:12 — 👍 5 🔁 3 💬 0 📌 0

Here’s the amazing Phylogenomics Group 2024 📸
Thank you to our incredible instructors for sharing their knowledge and making complex topics approachable and engaging! 🙌
And a big shoutout to all participants—Thanks for joining us this week🎉 @sabifo4.bsky.social @zihengyang.bsky.social
05.12.2024 11:53 — 👍 1 🔁 2 💬 0 📌 1

🚨 Exciting Day 3 Ahead! 🚨
Today, we’re honored to have one of the giants of Phylogenomics 🧬✨, @zihengyang.bsky.social, kicking off the day with two lectures on the Multispecies Coalescent Model.
Day3 will continue with an hands-on session led by Paschalia Kapli and Tomas Flouri🚀
04.12.2024 12:50 — 👍 3 🔁 1 💬 0 📌 0

✨ Day 2 highlight! ✨ Tomas Flouri and @sabifo4.bsky.social delivered masterful lectures and an exceptional hands-on session on Bayesian inference in Phylogenomics. 🎓💡 Their expertise truly shone through!
Excited for the insights Day 3 will bring—stay tuned! 🚀
04.12.2024 09:22 — 👍 1 🔁 2 💬 0 📌 0
The Physalia Phylogenomics course has started today! Tomas and Paschalia are leading today's session -- now, they are going through the practical session to apply all the concepts they have taught this morning! 💻
02.12.2024 15:13 — 👍 2 🔁 1 💬 0 📌 0
Last, but not least, thanks to @sabifo4.bsky.social for working on the PAML Wiki and for constantly supporting PAML users! 🙌 💫 Also, thanks to Janet Young and @sishuowang.bsky.social for their contributions to the PAML discussion group, which have been valuable for PAML users!🙌 (5/5)
29.11.2024 14:42 — 👍 3 🔁 0 💬 0 📌 0
If you experience technical issues when running PAML programs (e.g., programs abort, bugs, etc.), please raise an issue on the PAML GitHub repository. Please post all other questions on the PAML discussion group so that other PAML users can have their say and benefit from the discussion! 🌐 💬(4/n)
29.11.2024 14:32 — 👍 0 🔁 0 💬 1 📌 0
To post a message on the PAML discussion group, you must be part of the group. When requesting to join, please briefly introduce yourself and explain why you would like to join. This is a safety measure we have recently implemented to avoid bots disturbing the PAML community with SPAM. (3/n)
29.11.2024 14:29 — 👍 0 🔁 0 💬 1 📌 0
PAML discussion group - Google Groups
If you have gone through the Wiki and still have questions, please visit our PAML Discussion Group! 💻🌐 You can use the search bar to check whether someone has asked the same question before. If not, please share your question with the PAML community! (2/n)
groups.google.com/g/pamlsoftware
29.11.2024 14:27 — 👍 1 🔁 1 💬 1 📌 0
HOME
PAML is a program package for model fitting and phylogenetic tree reconstruction using DNA and protein sequence data. Please report only **technical issues** on this repository (e.g., compiling, pr...
If you have ever wondered whether the PAML documentation could be more interactive... We have good news for you! The PAML GitHub repository has been updated and a fresh PAML Wiki is now available for you to navigate the PAML documentation! 📄💻 (1/n)
github.com/abacus-gene/...
29.11.2024 14:26 — 👍 11 🔁 8 💬 1 📌 1