The best stories are the personal ones. So before we get to the new preprint, stay awhile and listen
#MobileLabs #TREC
24.02.2026 20:57 —
👍 21
🔁 9
💬 1
📌 1
Giant multicellular magnetotactic prokaryotes in marine sediments academic.oup.com/ismej/advanc... #jcampubs
17.02.2026 16:39 —
👍 39
🔁 11
💬 7
📌 2
Check out our latest pre-print of our favourite pets - the breviates. This work was led by Zhezhen Yi in collaboration with @andrewjroger.bsky.social and many others! By studying diverse breviates we were able to see how different metabolic pathways can evolved in response to low-oxygen.
09.02.2026 14:55 —
👍 23
🔁 14
💬 0
📌 0
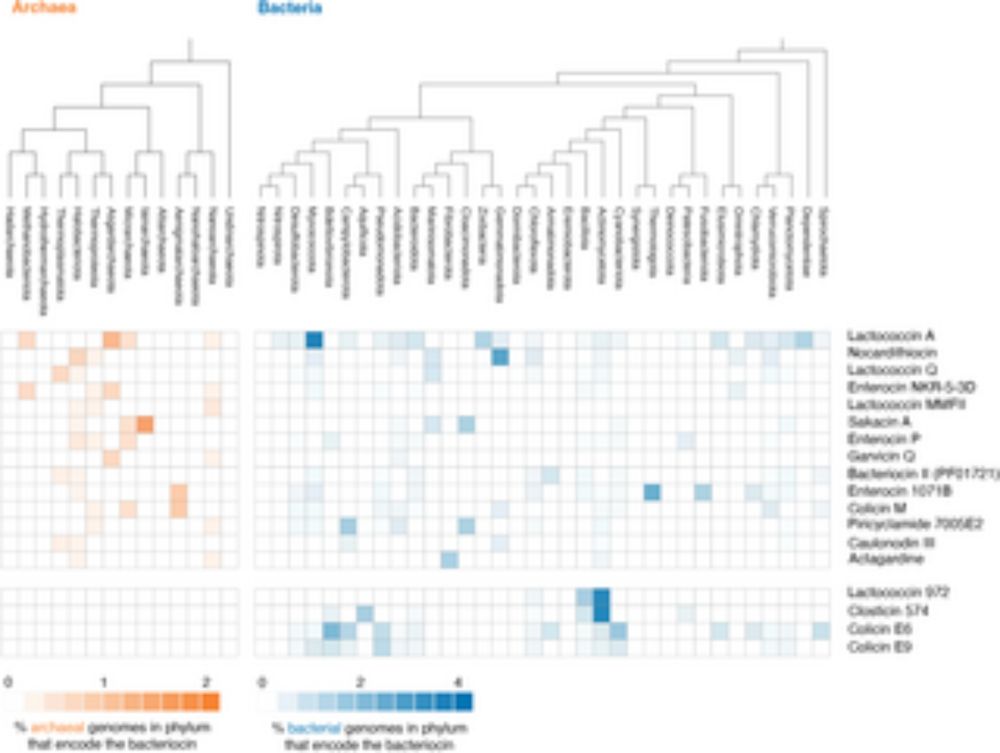
Archaea produce peptidoglycan hydrolases that kill bacteria
Archaea regularly interact with bacteria but reports of archaea killing bacteria are very rare. This study shows that many archaea encode peptidoglycan hydrolases, which specifically target bacterial ...
Archaea are often surrounded by bacteria. But is there ever active conflict between the two? Can archaea kill bacteria? If so, how do they do it?
Work by @romainstrock.bsky.social shows that some archaea can kill bacteria by secreting peptidoglycan hydrolases. journals.plos.org/plosbiology/...
14.08.2025 18:50 —
👍 145
🔁 65
💬 5
📌 5


Comparative Genomics of Unicellular Eukaryotes (San Feliu, Spain):
Abstract submission is open, with a short deadline! comparativegenomics2026.com
Join us as we explore the most diverse, surprising, and still largely uncharted branches of the eukaryotic tree.
02.02.2026 18:49 —
👍 20
🔁 12
💬 0
📌 1

My team at @cbitoulouse.bsky.social is recruiting a postdoc #bioinformatics with solid experience in metagenomic analyses.
Interest in evolution, ecology & MGEs is important.
The offer stands until the perfect candidate is found, and it could be you 🫵
🔁 🙏
#microSky #phagesky #UTIsky
@cnrs.fr
15.01.2026 11:19 —
👍 27
🔁 55
💬 1
📌 0

🚨 Postdoctoral Opportunity for Female Scientists🚨
The University of Vienna is awarding at least 20 fully funded 4 year postdoctoral positions to outstanding female scientists
Interested? Get in touch via direct message
careers.univie.ac.at/en/postdoc/e...
#MicroSky 🧪
#PostDoc @univie.ac.at
19.12.2025 00:24 —
👍 98
🔁 117
💬 0
📌 3
legend 2025 will be next Tuesday/Wednesday/Thursday.
The detailed program is on legend2025.sciencesconf.org?lang=en
If you are not attending the conference you can still follow the presentations online (Zoom links are on the website).
05.12.2025 11:45 —
👍 2
🔁 3
💬 0
📌 0
Oxygen production as an electron overflow pathway in ammonia-oxidizing archaea www.biorxiv.org/content/10.6... #jcampubs
04.12.2025 14:18 —
👍 10
🔁 8
💬 0
📌 0

The CNRS is breaking free from the Web of Science
From January 1st 2026, the CNRS will cut access to one of the largest commercial bibliometric databases, Clarivate Analytics'
From January 1st 2026, the CNRS will cut access to one of the largest commercial bibliometric databases, Clarivate Analytics' Web of Science, along with the Core Collection and Journal Citation Reports.
01.12.2025 11:04 —
👍 85
🔁 65
💬 0
📌 25

Exploring the human small intestinal luminal microbiome via a newly developed ingestible sampling device
Abstract. Because accessing the small intestine is technically challenging, studies of the small intestinal microbiome are predominantly conducted in patie
Exploring the human small intestinal luminal microbiome via a newly developed ingestible sampling device url: academic.oup.com/ismecommun/a....
I am proud to have contributed to this work with Alexandre Tronel, Thomas Soranzo (Pelican Health), @alegouellec.bsky.social + all other co-authors.
01.12.2025 20:47 —
👍 5
🔁 1
💬 0
📌 0

Une scientifique dit: "Attendez une minute... Les gens sur le spectre autistique sont sur-représentés dans la recherche scientifique. Mais... Mais ça veut dire que..."
Un autre réalise soudain, et murmure: "Mon Dieu..."
La légende dit: "L'autisme cause les vaccins."
Vaccines and Autism, par @zachweinersmith.bsky.social
15.11.2025 17:27 —
👍 529
🔁 70
💬 2
📌 3
So Nature-Springer, which absorbs several billion dollars in research funding each year, provides us with an API that attributes all citations to the first article in the volume.
Well done Springer Nature, at least I know where not to submit my next paper.
07.11.2025 17:15 —
👍 3
🔁 4
💬 0
📌 0

This paper has been a must! Great collaboration with @mkrupovic.bsky.social and @yifanzhou.bsky.social, a N&V by a legend of halophilic archaea tinyurl.com/yc3dcv72, and one picture of one of our expeditions to Dallol making the cover of the November issue of @natmicrobiol.nature.com
rdcu.be/eLtCH
02.11.2025 17:52 —
👍 78
🔁 32
💬 1
📌 2
Portail Emploi CNRS - Offre d'emploi - Postdoctoral researcher Microbiology (M/F)
JOB OFFER #PhageSky
We have a postdoc position opening in my group to investigate phage-MGE interactions !
We're based in the very nice city of Lyon, France @mmsb-lyon.bsky.social
Contact me for more info !
>> Apply on the CNRS webpage emploi.cnrs.fr/Offres/CDD/U...
22.10.2025 09:35 —
👍 29
🔁 42
💬 0
📌 0

The team at our research site in the alps
📣 New PhD student position opening in my lab!
Soils are full of microbes, but how many are active?
Surprisingly, we still lack reliable methods to answer this.
If you are interested in microbial dormancy and are fascinated by the Alps and glacier forefields, contact me.
🏔️ 🦠 💤 🧬 🎓
13.10.2025 12:20 —
👍 47
🔁 45
💬 2
📌 3
Portail Emploi CNRS - Offre d'emploi - Thèse en Microbiologie-Biochimie (H/F)
Open position to work on Type IX secretion (#T9SS) in our lab, in collaboration withe the group of Eric Reynolds at the Dental School of the University of Melbourne. Please spread the word, and forward to anyone potentially interested ! Apply here:
emploi.cnrs.fr/Offres/Docto...
09.10.2025 12:17 —
👍 25
🔁 35
💬 0
📌 2
![Exploring the space of self-reproducing RNA using generative models, Martin Weigt
Exploring the archaic introgression landscape of admixed populations through
joint ancestry inference, Jazeps Medina Tretmanis [et al.]
Predicting natural variation in the yeast phenotypic landscape with machine
learning, Sakshi Khaiwal [et al.]
Phylodynamic modeling with unsupervised Bayesian neural networks, Marino
Gabriele [et al.]
Likelihood-free inference of phylogenetic tree posterior distributions, Luc Blas-
sel [et al.]
Generative continuous time model reveals epistatic signatures in protein evolu-
tion, Barrat-Charlaix Pierre
Neural posterior estimation for high-dimensional genomic data from complex pop-
ulation genetic models, Jiseon Min [et al.]
A differentiable model for detecting diversifying selection directly from alignments
in large-scale bacterial datasets, Leonie Lorenz [et al.]
Detecting interspecific positive selection using transformers, Charlotte West [et al.]
Predicting Multiple Sequence Alignment Uncertainty via Machine Learning, Lucia
Martin-Fernandez [et al.]
Graph Neural Networks for Likelihood-Free Inference in Diversification Mod-
els, Amélie Leroy [et al.]
Popformer: learning general signatures of genetic variation and natural selection
with a self-supervised transformer, Leon Zong [et al.]](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:7x5c6ifrcznmg4mzhfuc2a6r/bafkreidur7bz43o2jhwr4gbt36awozawqthbkkfeszxjkxf4kvhyh7q6du@jpeg)
Exploring the space of self-reproducing RNA using generative models, Martin Weigt
Exploring the archaic introgression landscape of admixed populations through
joint ancestry inference, Jazeps Medina Tretmanis [et al.]
Predicting natural variation in the yeast phenotypic landscape with machine
learning, Sakshi Khaiwal [et al.]
Phylodynamic modeling with unsupervised Bayesian neural networks, Marino
Gabriele [et al.]
Likelihood-free inference of phylogenetic tree posterior distributions, Luc Blas-
sel [et al.]
Generative continuous time model reveals epistatic signatures in protein evolu-
tion, Barrat-Charlaix Pierre
Neural posterior estimation for high-dimensional genomic data from complex pop-
ulation genetic models, Jiseon Min [et al.]
A differentiable model for detecting diversifying selection directly from alignments
in large-scale bacterial datasets, Leonie Lorenz [et al.]
Detecting interspecific positive selection using transformers, Charlotte West [et al.]
Predicting Multiple Sequence Alignment Uncertainty via Machine Learning, Lucia
Martin-Fernandez [et al.]
Graph Neural Networks for Likelihood-Free Inference in Diversification Mod-
els, Amélie Leroy [et al.]
Popformer: learning general signatures of genetic variation and natural selection
with a self-supervised transformer, Leon Zong [et al.]
![PRIVET: PRIVacy metric based on Extreme value Theory, Antoine Szatkownik [et
al.]
Generative models for inferring the evolutionary history of the malaria vector
Anopheles gambiae, Amelia Eneli [et al.]
Language Models Outperform Supervised-Only Approaches for Conserved Ele-
ment Comprehension, Eyes Robson [et al.]
Identification and Classification of Orphan Genes, Spurious Orphan Genes, and
Conserved Genes from the human microbiome, Chen Chen
Neural Simulation-based inference of demography and selection, Francisco De
Borja Campuzano Jiménez [et al.]
Species Identification and aDNA Read Mapping Using k-mer Embeddings, Filip
Thor [et al.]
Contrastive Learning for Population Structure and Trait Prediction, Filip Thor [et
al.]
Protein and genomic language models chart a vast landscape of antiphage de-
fenses, Mordret Ernest
The Phylogenomics and Sparse Learning of Trait Innovations, Gaurav Diwan [et
al.]](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:7x5c6ifrcznmg4mzhfuc2a6r/bafkreig4akadduemzbit67bv7jnr7x2ircwiriykabg2imuf3phy6hhube@jpeg)
PRIVET: PRIVacy metric based on Extreme value Theory, Antoine Szatkownik [et
al.]
Generative models for inferring the evolutionary history of the malaria vector
Anopheles gambiae, Amelia Eneli [et al.]
Language Models Outperform Supervised-Only Approaches for Conserved Ele-
ment Comprehension, Eyes Robson [et al.]
Identification and Classification of Orphan Genes, Spurious Orphan Genes, and
Conserved Genes from the human microbiome, Chen Chen
Neural Simulation-based inference of demography and selection, Francisco De
Borja Campuzano Jiménez [et al.]
Species Identification and aDNA Read Mapping Using k-mer Embeddings, Filip
Thor [et al.]
Contrastive Learning for Population Structure and Trait Prediction, Filip Thor [et
al.]
Protein and genomic language models chart a vast landscape of antiphage de-
fenses, Mordret Ernest
The Phylogenomics and Sparse Learning of Trait Innovations, Gaurav Diwan [et
al.]
The decisions for LEGEND are out: legend2025.sciencesconf.org/data/book_le...
I'm really looking forward to hearing these 21 exciting presentations (and additional 30 posters) next December.
If you want to attend too, registration is open until October 17th through legend2025.sciencesconf.org
08.10.2025 11:04 —
👍 4
🔁 1
💬 1
📌 0
Job Details
Become my colleague @oxfordbiochemistry.bsky.social !
2 Associate Professorships in the areas of
- prokaryotic/eukaryotic microbiology
- metabolism
my.corehr.com/pls/uoxrecru...
29.09.2025 09:25 —
👍 30
🔁 43
💬 1
📌 1
We would like to thank the scientists across the world who characterize quinones in newly described species and publish their work e.g. in IJSEM from @microbiologysociety.org, Antonie van Leeuwenhoek, Archives of Microbiology and Systematic and Applied Microbiology. Quinones rock! 🤩 5/5
18.09.2025 10:10 —
👍 3
🔁 0
💬 0
📌 0
Validate User
...and after the study brilliantly conducted by Dr. Chobert on the dynamics of the quinone repertoire in Pseudomonadota that accompanied their metabolic diversification, also published earlier this year in the ISME Journal from @isme-microbes.bsky.social. 4/5
doi.org/10.1093/isme...
18.09.2025 10:10 —
👍 1
🔁 0
💬 1
📌 0

A novel quinone biosynthetic pathway illuminates the evolution of aerobic metabolism | PNAS
The dominant organisms in modern oxic ecosystems rely on respiratory quinones with
high redox potential (HPQs) for electron transport in aerobic re...
This much-needed compendium of quinones in bacteria is the last chapter of S. Chobert's PhD thesis. This comes after the discovery of a high potential quinone, the methylplastoquinone, led by @lipidorama.bsky.social and colleagues published earlier in @pnas.org... 3/5
doi.org/10.1073/pnas...
18.09.2025 10:10 —
👍 1
🔁 1
💬 1
📌 0
Among other discoveries, we show that the two menaquinone pathways tend to replace one another frequently, and that a lineage outside of Pseudomonadota unexpectedly show the potential to produce ubiquinone, a high redox potential usually found in Pseudomonadota and Eukaryota. 2/5
18.09.2025 10:10 —
👍 2
🔁 1
💬 1
📌 0
Really glad to announce a new preprint from our lab: Global distribution of quinones in Bacteria! We here combine comparative genomics and text mining to leverage information on quinones distribution from thousands of articles describing new species and from dozens of thousands of genomes! 1/5
18.09.2025 10:10 —
👍 12
🔁 5
💬 1
📌 1
Portail Emploi CNRS - Offre d'emploi - Ingénieur développeur - DevOps (H/F)
ATLASea #recrute:
Ingénieur développeur - DevOps (H/F) à RENNES
IT en contrat CDD de 13 mois
Date d'embauche prévue : 1 novembre 2025
Plus d'informations:
emploi.cnrs.fr/Offres/CDD/U...
#hiring #job
17.09.2025 14:02 —
👍 6
🔁 5
💬 0
📌 1















![Exploring the space of self-reproducing RNA using generative models, Martin Weigt
Exploring the archaic introgression landscape of admixed populations through
joint ancestry inference, Jazeps Medina Tretmanis [et al.]
Predicting natural variation in the yeast phenotypic landscape with machine
learning, Sakshi Khaiwal [et al.]
Phylodynamic modeling with unsupervised Bayesian neural networks, Marino
Gabriele [et al.]
Likelihood-free inference of phylogenetic tree posterior distributions, Luc Blas-
sel [et al.]
Generative continuous time model reveals epistatic signatures in protein evolu-
tion, Barrat-Charlaix Pierre
Neural posterior estimation for high-dimensional genomic data from complex pop-
ulation genetic models, Jiseon Min [et al.]
A differentiable model for detecting diversifying selection directly from alignments
in large-scale bacterial datasets, Leonie Lorenz [et al.]
Detecting interspecific positive selection using transformers, Charlotte West [et al.]
Predicting Multiple Sequence Alignment Uncertainty via Machine Learning, Lucia
Martin-Fernandez [et al.]
Graph Neural Networks for Likelihood-Free Inference in Diversification Mod-
els, Amélie Leroy [et al.]
Popformer: learning general signatures of genetic variation and natural selection
with a self-supervised transformer, Leon Zong [et al.]](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:7x5c6ifrcznmg4mzhfuc2a6r/bafkreidur7bz43o2jhwr4gbt36awozawqthbkkfeszxjkxf4kvhyh7q6du@jpeg)
![PRIVET: PRIVacy metric based on Extreme value Theory, Antoine Szatkownik [et
al.]
Generative models for inferring the evolutionary history of the malaria vector
Anopheles gambiae, Amelia Eneli [et al.]
Language Models Outperform Supervised-Only Approaches for Conserved Ele-
ment Comprehension, Eyes Robson [et al.]
Identification and Classification of Orphan Genes, Spurious Orphan Genes, and
Conserved Genes from the human microbiome, Chen Chen
Neural Simulation-based inference of demography and selection, Francisco De
Borja Campuzano Jiménez [et al.]
Species Identification and aDNA Read Mapping Using k-mer Embeddings, Filip
Thor [et al.]
Contrastive Learning for Population Structure and Trait Prediction, Filip Thor [et
al.]
Protein and genomic language models chart a vast landscape of antiphage de-
fenses, Mordret Ernest
The Phylogenomics and Sparse Learning of Trait Innovations, Gaurav Diwan [et
al.]](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:7x5c6ifrcznmg4mzhfuc2a6r/bafkreig4akadduemzbit67bv7jnr7x2ircwiriykabg2imuf3phy6hhube@jpeg)

