Yes, the way Cicero normalizes counts and computes pseudo cells from UMAP coordinates was really deleterious in our evaluation..
Maybe improvement will come with scATAC atlases, to compensate for data sparsity! 😊
30.09.2025 21:19 — 👍 1 🔁 0 💬 0 📌 0
Very happy to see this work finally online 🥳
It was really interesting to write a short manuscript mixing method presentation & broader insights on data preprocessing. 📝
Thank you to my co-supervisors @cantinilab.bsky.social & @juliosaezrod.bsky.social for their feedback on this side work! 🫶
5/5
30.09.2025 12:23 — 👍 3 🔁 3 💬 0 📌 1

CIRCE is a Python package based on #AnnData and fully compatible with the #scverse environment.
You can extract DNA region modules & visualize CIRCE's interactions over gene locations.
CIRCE also facilitates #CellOracle or #HuMMuS usage, making it unnecessary to run Cicero (code in R) first.
4/5
30.09.2025 12:23 — 👍 3 🔁 0 💬 1 📌 0

We explore the impact of preprocessing on DNA interactions inference. 👩🔬
We evaluated scATAC-seq preprocessings using promoter capture Hi-C data. 🧬
TLDR: Best performance came from single-cell inputs directly and from CIRCE metacells! 📊
In contrast, count normalization had a negative impact.
3/5
30.09.2025 12:23 — 👍 2 🔁 0 💬 2 📌 0

We compared CIRCE & Cicero on 2 datasets, demonstrating near identical results from the same input.
Both use pseudo-cells to reduce sparsity. CIRCE proposes a new strategy, whose output is closer to the single-cell profiles. 🛠️
On average, CIRCE uses 5x less memory and runs 150x faster ! 📈
2/5
30.09.2025 12:23 — 👍 1 🔁 0 💬 1 📌 0

I'm presenting #CIRCE, a Python package to infer co-accessible DNA region networks 🧬
Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️
30.09.2025 12:23 — 👍 4 🔁 3 💬 1 📌 0
I'm curious if other mid career PIs have considered completely abandoning the trad publication model. We've been making gradual moves towards that end goal. But it's been difficult to completely jump ship for one main reason for me at least. 1/
16.09.2025 05:43 — 👍 34 🔁 12 💬 8 📌 1

GitHub - cantinilab/scPRINT: single cell foundation model for Gene network inference and more
single cell foundation model for Gene network inference and more - cantinilab/scPRINT
On Tuesday at 2025-02-04 18:00 CET, @jkobject will talk about scPrint, a transformer model that infers gene networks from scRNA-seq data, at our 2nd community meeting of 2025! For more information, check out the GitHub: https://buff.ly/40RJ3gT & pre-print https://buff.ly/4hwK4Av
31.01.2025 18:02 — 👍 20 🔁 8 💬 2 📌 0
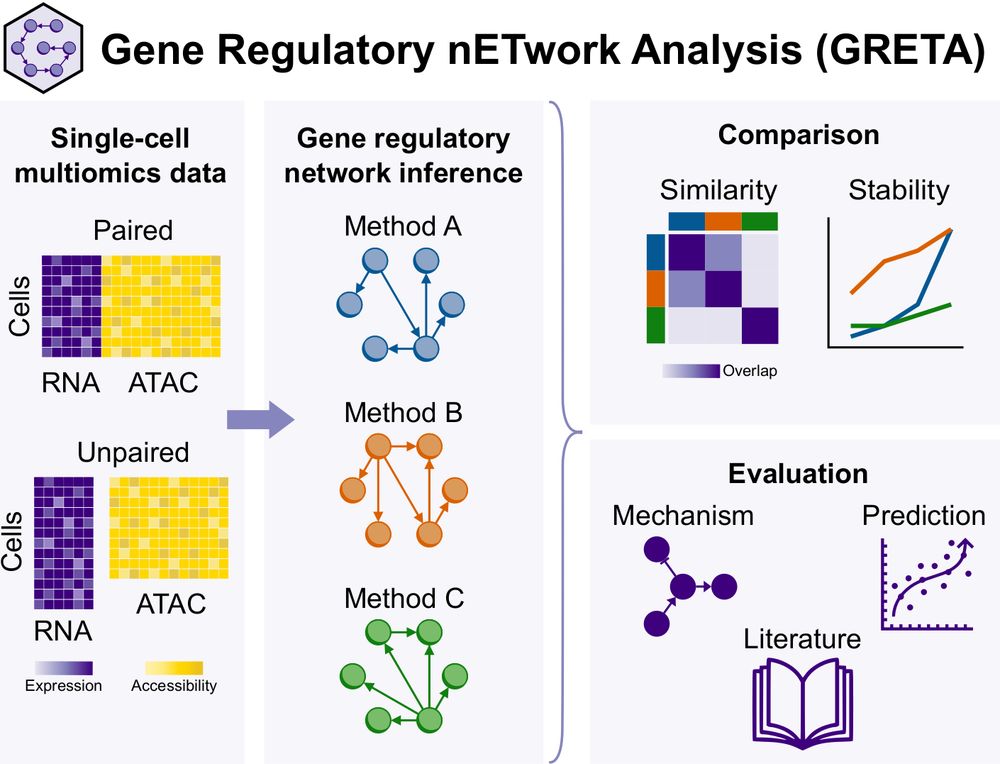
GRETA graphical abstract
We present Gene Regulatory nETwork Analsyis (GRETA), a framework to infer, compare and evaluate gene regulatory networks #GRNs. With it, we have benchmarked multimodal and unimodal GRN inference methods. Check the results here 👇
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
23.12.2024 08:43 — 👍 127 🔁 46 💬 1 📌 5
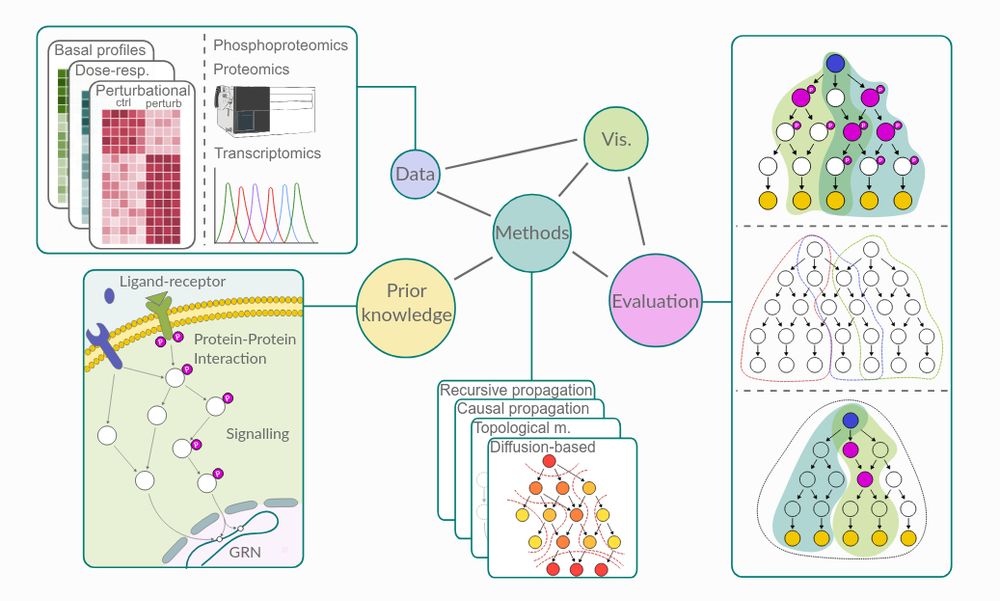
We present NetworkCommons, a unified platform 🪐 for network biology, providing access to omics data, knowledge, and contextualization methods, all with a consistent API 👇🧵
Paper: doi.org/10.1101/2024...
Docs: networkcommons.readthedocs.io
26.11.2024 09:44 — 👍 96 🔁 32 💬 2 📌 4
Congrats Pau for all this work !! 🎉🎉
I learned a lot from being around you (and I still do) 😇
And I'm really looking forward to seeing what you'll bring to the field in the coming years 🔮
25.11.2024 20:17 — 👍 1 🔁 0 💬 0 📌 0
I would love to be in too ! 😊
24.11.2024 21:59 — 👍 1 🔁 0 💬 0 📌 0

What are the key disrupted multicellular processes in heart failure? In our new work we combine 23 years of molecular data with recent single-cell atlases to draw a cross-study patient map
doi.org/10.1101/2024...
11.11.2024 09:06 — 👍 25 🔁 10 💬 0 📌 1
ORCID
Hi, would like to join the science feed too please ! :)
I'm an MD-PhD student currently working on single cell method development to predict molecular regulations.
orcid.org/0000-0001-87...
14.11.2024 06:18 — 👍 1 🔁 0 💬 0 📌 0
into brain evolution & development, open science, art & science https://www.youtube.com/watch?v=6hMNZHsrNHw, music, making, javascript, contemporary dance https://www.youtube.com/watch?v=OZfHj7F2FzQ
website: katjaq.github.io
Into brain development and evolution (+open science, generative art, genomics, music, 🇨🇱...). If you have questions about mechanical morphogenesis, I have more!
https://orcid.org/0000-0002-6671-858X
Yeast synthetic biology and trail running
Marie Skłodowska-Curie Actions postdoc in Spivakov Lab at Imperial College. Formerly postdoc at in Reilly Lab at Yale University. The sounds of noncoding variants wake me from my slumber.
Cancer Research Center of Toulouse and Barcelona Supercomputing Center. Interested in networks, heterogeneity and genome architecture. crct-inserm.fr/en/netbio2_en/
🎓 previously @borklab.bsky.social @EMBL.org & @saezlab.bsky.social @uniHeidelberg.bsky.social
📍Heidelberg
🦠Human microbiome | Host-microbiome interactions | Multiomics integration
Former CDC guy | Calm but Angry| He/him | Medical Science Communicator | Opinion =My own
🎓 PhD student in Biomedicine
📍 University of Helsinki, Finland
🧬 Ovarian cancer researcher
🏃♀️ Skier and runner
Head of Bioinformatics at Gordian Biotechnology. Open science proponent. Runner. 🧬🖥️🏃🏻
PhD candidate in @saezlab.bsky.social at the Institute for Computational Biomedicine, Heidelberg, Germany
phd student @saezlab.bsky.social 💻 ✨
trying to reduce pain in this world by studying diseases + drugs 💊
also super interested in figuring out how to prevent misinformation in society 🔑
Ph.D student in Computer Science and Engineering at the University of Washington working with Su-In Lee.
Teif lab at the University of Essex. We work on gene regulation in chromatin and applications to liquid biopsies, using approaches of genomics, biophysics, bioinformatics & AI. Our focus is nucleosomics, TF binding, CTCF, cfDNA. https://generegulation.org
Computational immunologist. Interested in immunooncology, cytometry, single cell data, spatial data, T and B cell specificities...
Professor of Biomedical Informatics @InfAtEd, Dir. @AI4BI_CDT - Bioinformatics, Machine Learning, BioNLP, Genomics, Genetic Disease
Berkeley professor (Bioeng, Compbio). Visiting Scientist at Calico. JBrowse genome browser / Apollo annotation editor, ML for gene regulation / molecular evolution / synbio. Occasional music, games, jokes
Principal Scientist @ Somite AI | Computational biology and biomedical data science | Boston, MA | https://mbernste.github.io








