
This is a joint work with @highlanderlab.bsky.social @gregorgorjanc.bsky.social
The software is available in github and will also be on CRAN as soon as the screening is cleared.
github.com/HighlanderLa...
@highlanderlab.bsky.social
Research on managing and improving populations at @RoslinInstitute & @TheDickVet. Led by the chief Highlander @GregorGorjanc.

This is a joint work with @highlanderlab.bsky.social @gregorgorjanc.bsky.social
The software is available in github and will also be on CRAN as soon as the screening is cleared.
github.com/HighlanderLa...

Lastly I gave a short course on Stochastic simulations of breeding programmes with AlphaSimR - a teaser for our free on-line course www.edx.org/learn/animal...
03.02.2025 10:57 — 👍 1 🔁 1 💬 0 📌 0I presented the work of Letícia Lara on Evaluation of selective breeding programme designs for black soldier fly larvae body weight (with María Martínez Castillero, Thiago Oliveira, Ivan Pocrnić, Jana Obšteter)
03.02.2025 10:57 — 👍 1 🔁 1 💬 1 📌 0Jana Obšteter presented the work of Laura Strachan on Optimizing pedigree reconstruction and patriline determination in honeybees (with Jernej Bubnič and Janez Presern)
03.02.2025 10:57 — 👍 1 🔁 1 💬 1 📌 0Irene de Carlos Fernández presented her work on Modelling the Impact of Non-Native Honey Bee Importation on Native Apis mellifera mellifera Populations (with Laura Strachan, Grace McCormack, Jana Obšteter)
03.02.2025 10:57 — 👍 1 🔁 1 💬 1 📌 0Last week some of us attended the 1st EAAP - European Federation of Animal Science Insect Genetics Workshop, which was organised with InsectIMP CA22140 COST Action action by Gertje Petersen and Jana Obšteter. We presented
03.02.2025 10:57 — 👍 1 🔁 1 💬 1 📌 0@h-a-n.bsky.social
17.01.2025 06:19 — 👍 0 🔁 0 💬 0 📌 0
Xinger (Evie) Tang presented a poster “A Pedigree-Based Method for Localization of Recombination Events”
17.01.2025 06:15 — 👍 2 🔁 1 💬 0 📌 0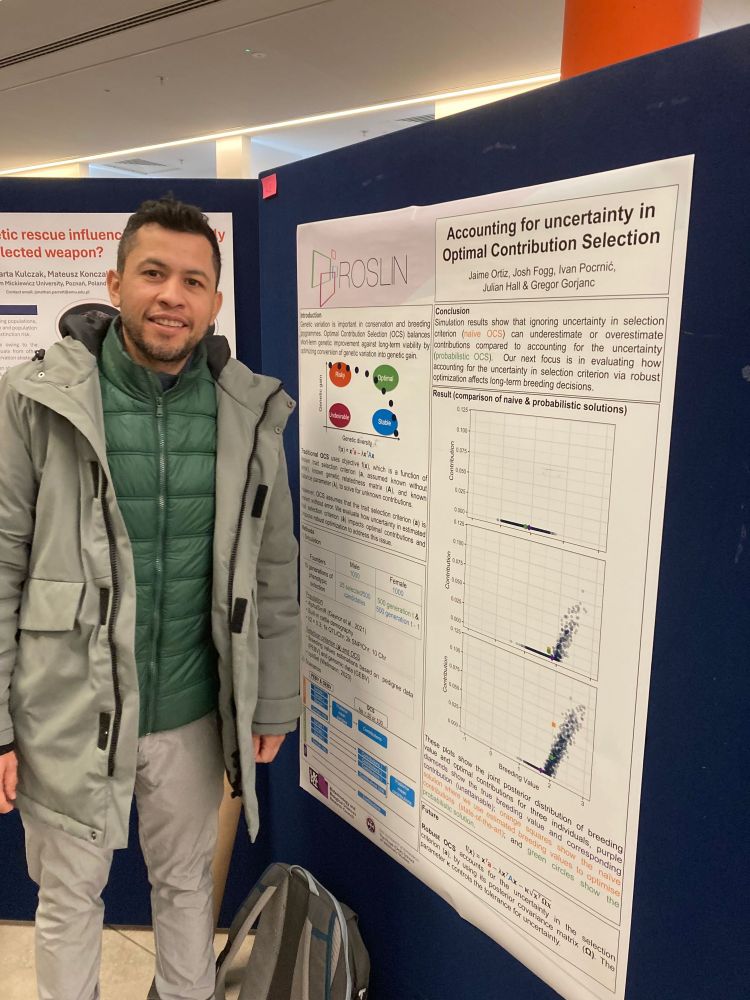
Jaime Ortiz Cuadros presented a poster “Accounting for uncertainty in Optimal Contribution Selection” - joint work with Josh Fogg, Julian Hall, and Ivan Pocrnic
17.01.2025 06:15 — 👍 1 🔁 1 💬 1 📌 0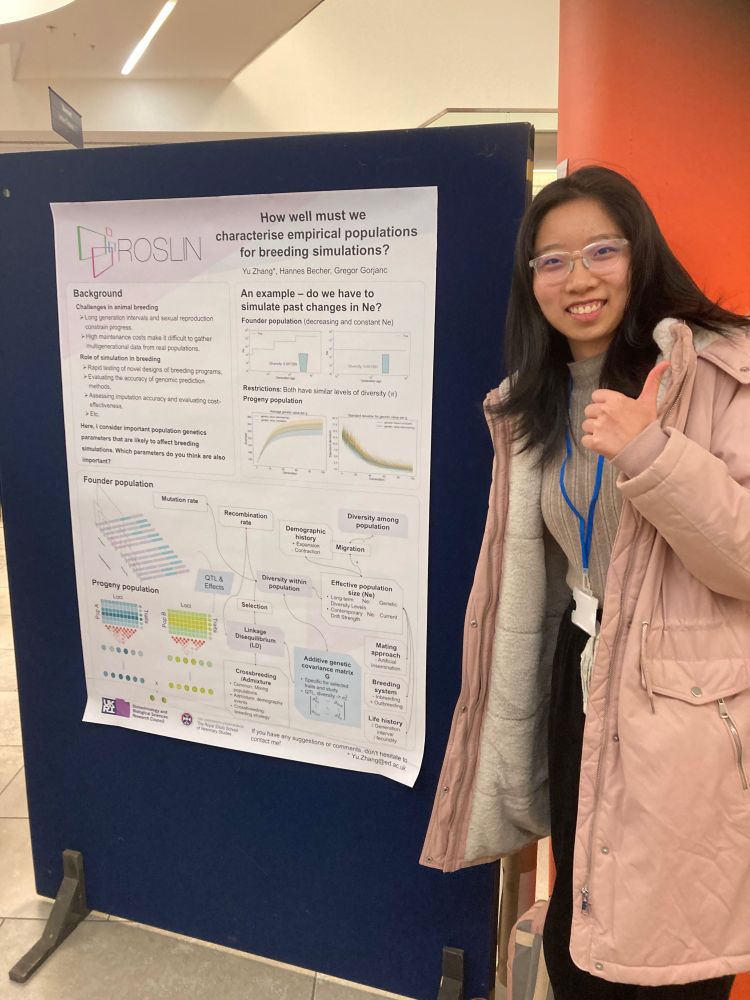
Yu (Uni) Zhang presented a poster “How well must we characterise empirical populations for breeding simulations?”
17.01.2025 06:15 — 👍 1 🔁 1 💬 1 📌 0
Hannes Becher presented a poster “Fast pedigree PCA with the randPedPCA R package”- joint work with @epigenci.bsky.social
17.01.2025 06:15 — 👍 2 🔁 1 💬 1 📌 1
Ros Craddock talked about “General Pedigree Tracking of Disease-causing Alleles for Recessive Monogenic Diseases in Dogs” - joint work with Joanna ilska, Cathryn Mellersh, Pam Wiener, and Audrey Martin
17.01.2025 06:15 — 👍 1 🔁 1 💬 1 📌 0Gabriela Mafra Fortuna talked abou “Experience with Inferring Ancestral Recombination Graph of the Global Cattle Population” - joint work with Jana Obsteter
17.01.2025 06:15 — 👍 1 🔁 1 💬 1 📌 0
HighlanderLab at PopGroup58 in Sheffield! It’s always interesting to attend the highly diverse set of talks and posters on population genetics and associated fields at the PopGroup conference. This year it was hosted by the University of Sheffield. We contributed with two talks and four posters.
17.01.2025 06:15 — 👍 2 🔁 1 💬 2 📌 0
Exciting PhD opportunity to join us at Roslin/Edinburgh and work with the largest whole-genome sequencing dataset in selective breeding. www.findaphd.com/phds/project...
29.11.2024 10:12 — 👍 7 🔁 9 💬 0 📌 1
A new pre-print “Robust Optimal Contribution Selection” led by Josh Fogg. We show how to account for the uncertainty and correlation of estimated breeding values in the optimal contribution selection problem: arxiv.org/abs/2412.02888
10.12.2024 07:40 — 👍 1 🔁 2 💬 2 📌 0AlphaSimR course edin.ac/3wfGSEj Week 2:
It’s time for trait simulation - one of the key reasons we developed AlphaSimR - to study the principles of quantitative genetics and selective breeding. Here, we show the outline of such a simulation, ...

📺 In our seminar last week Dr Hannes Becher from
@highlanderlab.bsky.social spoke about "Assembly-free estimation of population parameters using single-individual k-mer spectra". 🧬💻
If you missed the seminar, the recording is now available on our YouTube channel 👇
youtu.be/-wHqNbdpEog?...
🧪

Please share: Exciting post for a Quantitative Genetics and Breeding scientist to
to develop and implement a breeding programme for artemia - a Knowledge Transfer Partnership between Aquanzo and Roslin Institute supported by KTN/KTP InnovateUK elxw.fa.em3.oraclecloud.com/hcmUI/Candid...
AlphaSimR course edin.ac/3wfGSEj Week 2: Continuing work with genomes and genotype variation, focusing on loci. We extract genotypes from a population, calculate and plot allele frequencies, and calculate and plot linkage-disequilibrium.
11.03.2024 10:13 — 👍 3 🔁 3 💬 1 📌 1We thank the support from @roslininstitute @TheDickVet @EdinburghUni @BBSRC @TRAINatEd_MSCA @EdinInnovations @Bayer4Crops @UniNewEngland. Fin.
12.02.2024 04:38 — 👍 0 🔁 0 💬 0 📌 0
All course material is available at: jvanderw.une.edu.au/aabc2024.htm - enjoy your independent study;)
12.02.2024 04:37 — 👍 0 🔁 0 💬 1 📌 0Day 5 returned to topics of Day 1 by diving into Ancestral Recombination Graphs (ARGs), coalescent simulations, and fitting linear mixed models on tree structures, including our current experiences in working with ARGs in agricultural datasets.
12.02.2024 04:37 — 👍 0 🔁 0 💬 1 📌 0This part used FieldSimR available at cran.r-project.org/package=Fiel... and GxE simulations desribed in doi.org/10.21203/rs....
12.02.2024 04:37 — 👍 0 🔁 0 💬 1 📌 0
Day 4 dived into plant breeding trials, experimental design, spatial variation, genotype by environment (GxE) interactions, and how to simulate and model these sources of variation.
12.02.2024 04:37 — 👍 0 🔁 0 💬 1 📌 0
Day 3 reversed the flow from forward data simulations to inverse probability statements about unknown parameters from the observed data - all based on linear mixed models with pedigree and genomic data.
12.02.2024 04:36 — 👍 0 🔁 0 💬 1 📌 0

Day 2 delved into quantitative genetics, how it is implemented in AlphaSimR, and demonstrating fundamental results from the theory. With AlphaSimR we can now easily inspect where and how these fundamental results come about!
12.02.2024 04:36 — 👍 0 🔁 0 💬 1 📌 0Showcase of plant breeding simulations is available at www.biorxiv.org/content/10.1.... Animal breeding showcase is in development.
12.02.2024 04:35 — 👍 0 🔁 0 💬 1 📌 0
AlphaSimR is available from cran.r-project.org/package=Alph...
Free intro on-line course on AlphaSimR is available from www.edx.org/course/breed...


Day 1 started with an overview of simulating breeding programmes, the essential components of such simulations, and AlphaSimR implementation with simple and advanced examples.
12.02.2024 04:34 — 👍 0 🔁 0 💬 1 📌 0