My latest in @plosbiology.org Look I get it, it seems really dark, but there are opportunities. My paper explores some ideas and tries to guide ways of thinking through the anticipated challenges. An emphasis on America’s “biohubs” and entrepreneurship
journals.plos.org/plosbiology/...
13.04.2025 14:04 — 👍 164 🔁 45 💬 2 📌 2

An illustration of the candidate vaccine against Clostridioides difficile infection.
A recent study of an mRNA vaccine in a mouse model of 𝘊𝘭𝘰𝘴𝘵𝘳𝘪𝘥𝘪𝘰𝘪𝘥𝘦𝘴 𝘥𝘪𝘧𝘧𝘪𝘤𝘪𝘭𝘦 infection provided proof of concept of a protective effect. Learn more: nej.md/4kS8x5X
@umich.edu #MedSky #IDSky
02.04.2025 18:03 — 👍 23 🔁 9 💬 0 📌 3
Here's my interview with @stevenstrogatz.com on @quantamagazine.bsky.social's Joy of Why podcast.
This was *so much fun*. I was nervous going into it (I really look up to Steve!), but I had a blast and I think this is the best interview I have ever done.
Thanks for having me on, folks!
20.03.2025 16:14 — 👍 97 🔁 17 💬 5 📌 3
PNAS
Proceedings of the National Academy of Sciences (PNAS), a peer reviewed journal of the National Academy of Sciences (NAS) - an authoritative source of high-impact, original research that broadly spans...
I’m thrilled to share my first ever publication, now published in PNAS! www.pnas.org/doi/10.1073/...
With mentorship from the amazing @ksxue.bsky.social, I looked at how the outcomes of species introductions to microbial communities are influenced by the number of introduced microbes.
11.03.2025 13:22 — 👍 89 🔁 31 💬 1 📌 1
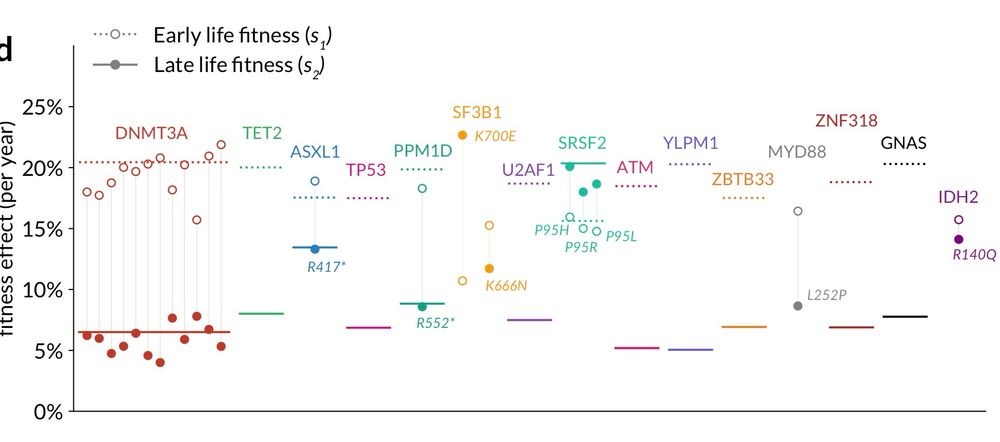
Delighted to share a major update on our work investigating age-related deceleration in clonal haematopoiesis.
Takehome: Widespread and substantial deceleration in fitness with age!
Amazing effort by PhD student Hamish MacGregor 💪
www.biorxiv.org/content/10.1...
06.03.2025 19:48 — 👍 15 🔁 7 💬 1 📌 1

Artist's depiction of mutant lineages rising and falling in frequency, influenced by selection, recombination, and genetic drift. In this issue, Lyulina et al. develop a mathematical framework for quantifying the statistical associations between these variants. Image courtesy of Anastasia Lyulina.
I am glad to see this work in print & on the cover! Read more at academic.oup.com/genetics/article/228/3/iyae145/7747749
06.11.2024 19:02 — 👍 8 🔁 1 💬 0 📌 0
Fascinating work showing genome-wide DNA packaging into phage capsids in "normie" gut bacteria!
27.11.2024 00:33 — 👍 5 🔁 1 💬 0 📌 0
Really cool and clever study! We saw extensive core genome recombination in gut bacteria in recent work, and I’ve always wondered about the underlying mechanism -- phage-like elements seem like the best candidate. Your examples of LT and GTA are particularly striking.
27.11.2024 00:25 — 👍 0 🔁 0 💬 0 📌 0

Asynchronous abundance fluctuations can drive giant genotype frequency fluctuations
Nature Ecology & Evolution - Based on a combination of experiments and modelling, this study shows large stochastic fluctuations in genotype frequencies caused by intrinsic and extrinsic...
We usually think of genetic drift as the predominant stochastic force in evolving populations. But working with some model microbial populations, we found a distinct source of demographic stochasticity that scales (and behaves) differently than drift
Learn more in our new paper 👉 rdcu.be/d07Np
22.11.2024 15:46 — 👍 28 🔁 10 💬 2 📌 1
Xue lab at UC Irvine
Ecology and evolution in the human gut microbiome
Hi friends new and old! I study how microbes interact and evolve in complex communities like the human gut microbiome.🦠🧬💩 I'm thrilled to share that I'm starting a lab at UC Irvine in April 2025 and am recruiting at all levels - please spread the word! kxuelab.com More about my work below...🧵1/n
19.11.2024 18:36 — 👍 390 🔁 115 💬 24 📌 10
Releases · cooplab/popgen-notes
Population genetics notes. Contribute to cooplab/popgen-notes development by creating an account on GitHub.
Just posting this to #popgen
Here's a link to my notes on population & quantitative genetics:
github.com/cooplab/popg...
Hoping to extend it more after the winter holidays, as I'm just finishing up teaching the undergrad version of class.
19.11.2024 23:37 — 👍 362 🔁 159 💬 12 📌 1

Somatic mutation and selection at epidemiological scale
As we age, many tissues become colonised by microscopic clones carrying somatic driver mutations ([1][1]–[10][2]. Some of these clones represent a first step towards cancer whereas others may contribu...
Resharing here a recent X post. In this preprint, we introduce an improved version of NanoSeq, a duplex sequencing protocol with <5 errors per billion bp in single DNA molecules, and use it to study the somatic mutation landscape of oral epithelium in >1000 people. 1/ www.medrxiv.org/content/10.1...
20.11.2024 14:27 — 👍 88 🔁 38 💬 2 📌 5
My first theory project! I've really enjoyed how we could decompose complex dynamics into those of individual lineages.
This approach is quite flexible, and we're hoping to extend it to more complex scenarios, including N(t) and certain forms of positive selection!
23.04.2024 19:20 — 👍 7 🔁 2 💬 0 📌 0
Thanks for the kind words☺️
24.02.2024 18:18 — 👍 1 🔁 0 💬 0 📌 0
PS: after a long revision process, this paper has gotten much better but admittedly a bit long (with 41 SI figures 🙂). So don't hesitate to reach out if you have any questions!
See also Ben's thread on twitter when we first posted on biorxiv
x.com/benjaminhgoo...
(n/n)
14.02.2024 00:20 — 👍 4 🔁 0 💬 1 📌 0

and 2) global spread of particular sequences. (btw @rwolff.bsky.social and @nanditagarud.bsky.social recently found more evidence of global selective sweeps in these gut bacteria. www.biorxiv.org/content/10.1...)
14.02.2024 00:18 — 👍 4 🔁 0 💬 1 📌 0

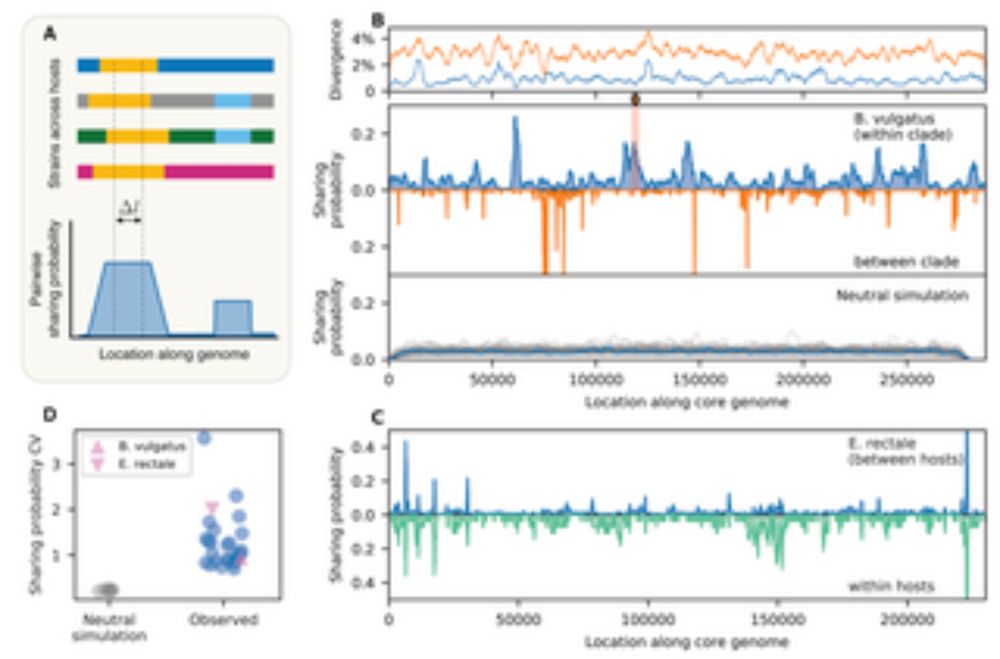
We also found some evidence that many recombined segments are strongly selected, leading to 1) within-host sweeps
14.02.2024 00:15 — 👍 2 🔁 0 💬 1 📌 0

Fig W buried in SI, showing limited role of sequence divergence in these gut species
What might cause all these fine-scale variations? Many factors are commonly thought to impact realized recombination rate. Here we were able to rule out the effect of sequence divergence, which plays very little role in almost all gut species (Fig 4 & W in SI are probably my favorite figures)
14.02.2024 00:12 — 👍 2 🔁 0 💬 1 📌 0

We found extensive heterogeneity in the rates and lengths of recombination, both between species and within species.
14.02.2024 00:09 — 👍 3 🔁 0 💬 2 📌 0

However, that was only a coarse summary of recombination at the species level. Using closely related strains found in the dataset, we were able to directly identify individual recombination events.
14.02.2024 00:08 — 👍 2 🔁 0 💬 1 📌 0

fig 1F
Here are some highlights of what we found. First and foremost, gut bacterial species recombine quite a lot -- certainly closer to H pylori than to M tuberculosis, the two extremes in the microbial world when it comes to recombination
14.02.2024 00:07 — 👍 2 🔁 0 💬 1 📌 0
For example, do gut bacteria recombine on human-relevant timescales?
Do recombination events even matter for evolution (i.e. get selected), since it's only mixing existing genes?
What are the mechanisms responsible for this recombination?
14.02.2024 00:05 — 👍 2 🔁 0 💬 1 📌 0

Ecology drives a global network of gene exchange connecting the human microbiome - Nature
Horizontal gene transfer — the exchange of genetic material between different species or lineages — is an important factor in bacterial evolution. A study of human microbiome data comprising more than...
So, there is a ton of work about HGT between species and how mobile elements hop around in this ecosystem. But genetic exchange also mixes up the core genome within a species, enabling bacteria to *recombine* like sexual organisms. Much less is known about recombination in gut bacteria:
14.02.2024 00:05 — 👍 2 🔁 0 💬 1 📌 0
Using metagenomic data, we quantified the dynamics of this process both at the scale of a single human host and across the global population.
14.02.2024 00:03 — 👍 3 🔁 0 💬 1 📌 0

Bacterial ecology and evolution converge on seasonal and decadal scales
Ecology and evolution are distinct theories, but the short lifespans and large population sizes of microbes allow evolution to unfold along contemporary ecological time scales. To document this in a natural system, we collected a two-decade, 471-metagenome time series from a single site in a freshwater lake, which we refer to as the TYMEFLIES dataset. This massive sampling and sequencing effort resulted in the reconstruction of 30,389 metagenomic-assembled genomes (MAGs) over 50% complete, which dereplicated into 2,855 distinct genomes (>96% nucleotide sequence identity). We found both ecological and evolutionary processes occurred at seasonal time scales. There were recurring annual patterns at the species level in abundances, nucleotide diversities (π), and single nucleotide variant (SNV) profiles for the majority of all taxa. During annual blooms, we observed both higher and lower nucleotide diversity, indicating that both ecological differentiation and competition drove evolutionary dynamics. Overlayed upon seasonal patterns, we observed long-term change in 20% of the species' SNV profiles including gradual changes, step changes, and disturbances followed by resilience. Most abrupt changes occurred in a single species, suggesting evolutionary drivers are highly specific. Nevertheless, seven members of the abundant Nanopelagicaceae family experienced abrupt change in 2012, an unusually hot and dry year. This shift coincided with increased numbers of genes under selection involved in amino acid and nucleic acid metabolism, suggesting fundamental organic nitrogen compounds drive strain differentiation in the most globally abundant freshwater family. Overall, we observed seasonal and decadal trends in both interspecific ecological and intraspecific evolutionary processes. The convergence of microbial ecology and evolution on the same time scales demonstrates that understanding microbiomes requires a new unified approach that views ecology and evolution as a single continuum. ### Competing Interest Statement The authors have declared no competing interest.
Been dreaming of this paper for a decade. 1 PhD and 1 postdoc later, here it is!
What do ecology and evolution look like in a 20-year microbiome time series? They blur together
@quendi.bsky.social @archaeal.bsky.social @uslter.bsky.social @sarilog.bsky.social 🧪🖥️🧬
www.biorxiv.org/content/10.1...
08.02.2024 23:47 — 👍 43 🔁 24 💬 5 📌 1

Changing fitness effects of mutations through long-term bacterial evolution
Predictable and parallel changes occur in the fitness effects of mutations in Escherichia coli over 50,000 generations.
How stable are bacterial genomes as they adapt to an environment? My collaboration with
Anurag Limdi, Alex Couce, @relenski.bsky.social, and
Olivier Tenaillon exploring this over 50,000 generations of evolution is out today! 1/
(cross-post from the other place)
www.science.org/doi/10.1126/...
26.01.2024 20:27 — 👍 100 🔁 51 💬 1 📌 4
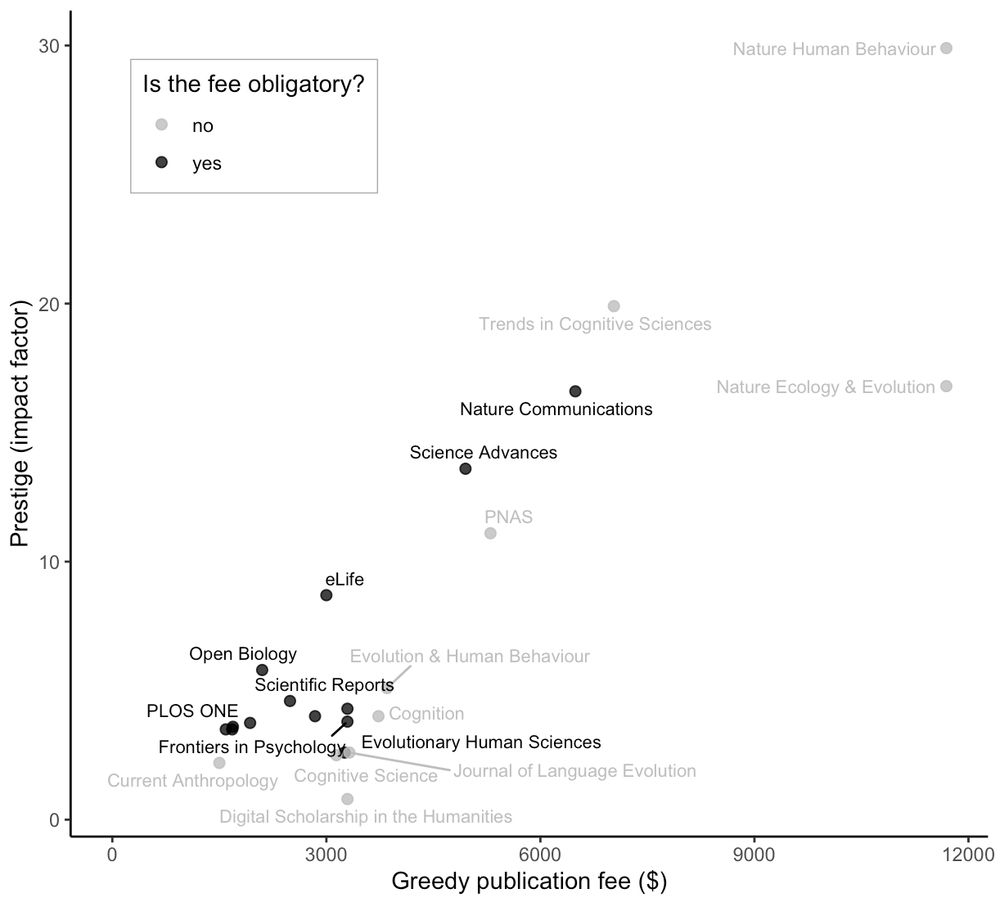
A plot showing prestige (impact factor) on vertical axis and fees charged by journals on horizontal axis.
Journal fees for open access are becoming obscene. But what are we, scientists, paying for? I made a simple plot with journals in my research area(s). Clearly, we are paying for prestige: a shockingly clean correlation between the impact factor and journal fees
13.12.2023 13:06 — 👍 303 🔁 180 💬 21 📌 19
Group leader Research Institute of Molecular Pathology (IMP) impvienna.bsky.social in Vienna, trained at MSKCC, working on #cancer #tumorimmunology #resistance. Mentor. Mum. Views are my own.
https://www.obenauflab.com/
https://www.imp.ac.at/
Max Planck Research Group Leader // Plant pathogen emergence and evolution // http://instagram.com/honourmccann // Opinions my own etc
Machine Learning & Systems Biology. ML Group Leader @arcinstitute. PhD @StanfordAILab
http://www.yusufroohani.com
Ph.D. candidate in Biology at Stanford University - speciation, conservation, and population genomics
@embl.org PhD & Bridging Excellence Postdoc (LifeScienceAlliance) with
@petrovadmitri.bsky.social @stanford.edu and Aulehla & Steinmetz labs @embl.org using #SystemsEvolution to study #Aging
mexican in Germany with an open love for tacos
Associate Editor at #PLOSBiology for #openaccess, working with #microbiology and #immunology
https://scheduler.zoom.us/melissa-vazquez-hernandez/meet-the-editor-melissa
Views are my own
MRC Career Development Award fellow at the University of Cambridge | Group Leader of the Microbiome Function and Diversity lab (https://microfundiv-lab.github.io) | Bioinformatics, Machine Learning, Metagenomics, Functional modelling, Human microbiome
Department of Host-Microbe Interactions, St. Jude Children’s Research Hospital
Memphis, TN
TCRs, influenza virus, anti-tumor immunity, books, dogs, Venice
Editor in Chief of the #NonProfit, #OpenAccess journal @plosbiology.org Former Chief Editor of Nature Microbiology.
#Virologist. #Feminist. #Spaniard in the UK. #Galician. #European always.
Views my own.
https://orcid.org/0000-0002-3666-5683
Group leader at I2BC | EVOGEN team | Evolutionary geneticist interested in intragenomic conflicts and genome evolution.
PhD Candidate in the Pritchard Lab at Stanford University. Interested in statistical and population genetics.
https://nikhilmilind.dev/
assistant professor at ucsf interested in genetics, statistics, etc…
jeffspence.github.io
Immunologist. T cell exhaustion, immunotherapy, Immune Health.
Postdoctoral scientist @ Sanger Institute in Cambridge, UK. Sperm sequencing, de novo mutation, and somatic evolution.
Group Leader @Wellcome Sanger Institute. @CRUK fellow | Interested in mutations acquired during aging and their impact across generation.
Professor of EECS and Statistics at UC Berkeley. Mathematical and computational biologist.
Evolution, cancer, immunology, math(s)
Ursula Zoellner Professor of Cancer Research
University of Cambridge
blundelllab.com
Scientist. Group leader at the Sanger Institute, Cambridge UK. Somatic mutation and selection in normal tissues, cancer and ageing.
Mobile genetic element whisperer. Computational Biologist @arcinstitute.org. Genetics PhD @stanfordmedicine.bsky.social.
I post about programmable recombinases and genome design.